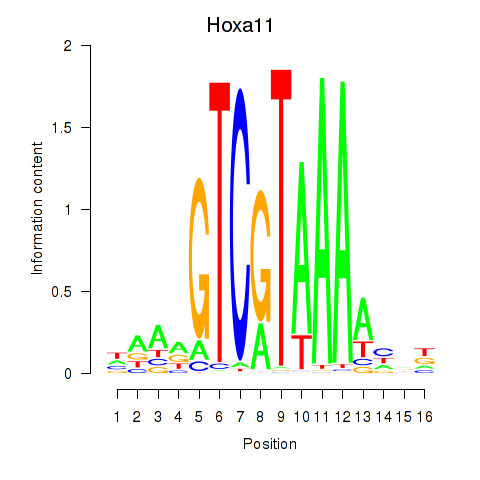
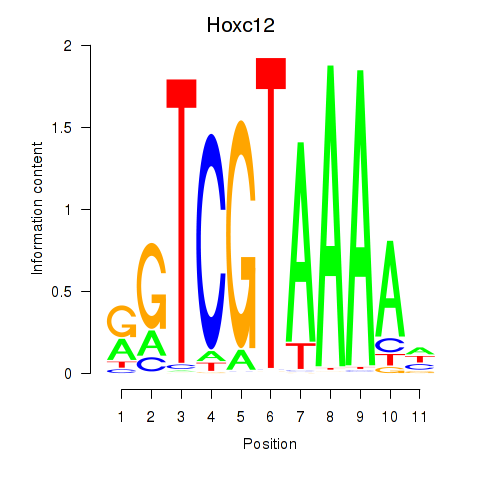
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hoxa11_Hoxc12
Z-value: 0.54
Transcription factors associated with Hoxa11_Hoxc12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa11
|
ENSRNOG00000059870 | homeobox A11 |
|
Hoxa11
|
ENSRNOG00000053884 | homeobox A11 |
|
Hoxc12
|
ENSRNOG00000016116 | homeo box C12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa11 | rn6_v1_chr4_-_82215022_82215022 | 0.24 | 2.0e-05 | Click! |
| Hoxc12 | rn6_v1_chr7_+_144547939_144547939 | 0.08 | 1.3e-01 | Click! |
Activity profile of Hoxa11_Hoxc12 motif
Sorted Z-values of Hoxa11_Hoxc12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_152533349 | 25.23 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr4_+_14151343 | 18.36 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr1_+_201055644 | 16.97 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_-_73958480 | 15.60 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr8_-_55087832 | 14.69 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr3_+_53563194 | 13.64 |
ENSRNOT00000048300
|
Xirp2
|
xin actin-binding repeat containing 2 |
| chr4_-_159192526 | 10.94 |
ENSRNOT00000026731
|
Kcna1
|
potassium voltage-gated channel subfamily A member 1 |
| chr19_-_22194740 | 10.02 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr4_-_82209933 | 9.43 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chrX_+_121612952 | 9.32 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr10_-_104179523 | 9.19 |
ENSRNOT00000005292
|
Slc25a19
|
solute carrier family 25 member 19 |
| chr4_-_82295470 | 9.17 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr1_+_8310577 | 7.24 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr4_+_14071507 | 7.22 |
ENSRNOT00000066224
ENSRNOT00000075962 |
Cd36
RGD1565355
|
CD36 molecule similar to fatty acid translocase/CD36 |
| chr2_-_18565842 | 7.18 |
ENSRNOT00000067456
ENSRNOT00000063821 ENSRNOT00000045532 |
Vcan
|
versican |
| chrX_+_131617798 | 6.88 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr1_-_93949187 | 6.79 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr13_-_51297621 | 6.41 |
ENSRNOT00000030926
|
Ndufv3
|
NADH:ubiquinone oxidoreductase subunit V3 |
| chr5_-_154332940 | 6.36 |
ENSRNOT00000014458
|
Pithd1
|
PITH domain containing 1 |
| chr16_+_81587446 | 5.83 |
ENSRNOT00000092680
|
NEWGENE_1582994
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr4_-_82202096 | 5.56 |
ENSRNOT00000081824
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chrX_+_42951237 | 5.36 |
ENSRNOT00000060113
|
RGD1559536
|
similar to vitellogenin-like 1 precursor |
| chr1_-_211265161 | 5.32 |
ENSRNOT00000080041
ENSRNOT00000023477 |
Bnip3
|
BCL2 interacting protein 3 |
| chr1_-_53038229 | 5.15 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr4_-_34194764 | 5.10 |
ENSRNOT00000045270
|
Col28a1
|
collagen type XXVIII alpha 1 chain |
| chr10_+_14543200 | 4.46 |
ENSRNOT00000047111
|
RGD1565784
|
RGD1565784 |
| chr9_+_50526811 | 4.46 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr1_+_94718402 | 4.44 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr7_-_11963268 | 4.25 |
ENSRNOT00000080342
ENSRNOT00000025266 |
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr1_-_207811008 | 3.62 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr6_+_18970564 | 3.59 |
ENSRNOT00000090121
ENSRNOT00000030803 |
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr1_-_169721758 | 3.49 |
ENSRNOT00000023213
|
LOC100910107
|
ubiquitin carboxyl-terminal hydrolase DUB-1-like |
| chrX_+_110007214 | 3.32 |
ENSRNOT00000093539
|
Nrk
|
Nik related kinase |
| chr9_+_64745051 | 3.24 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr5_-_156734541 | 3.19 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr11_-_61470046 | 3.17 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr11_-_4397361 | 3.11 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr17_+_77195247 | 3.08 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr4_-_56493923 | 2.61 |
ENSRNOT00000027173
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr10_-_88009576 | 2.55 |
ENSRNOT00000018566
|
Krt35
|
keratin 35 |
| chr2_+_230901126 | 2.44 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr16_-_20511818 | 2.37 |
ENSRNOT00000026567
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_122019301 | 2.35 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr6_-_105160470 | 2.28 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr5_+_154489590 | 2.21 |
ENSRNOT00000035788
|
Id3
|
inhibitor of DNA binding 3, HLH protein |
| chr11_+_60371729 | 2.20 |
ENSRNOT00000058453
|
Cd200
|
Cd200 molecule |
| chr9_-_69960086 | 2.16 |
ENSRNOT00000072737
|
Gpr1
|
G protein-coupled receptor 1 |
| chr10_-_29196563 | 2.15 |
ENSRNOT00000005205
|
Fabp6
|
fatty acid binding protein 6 |
| chr7_-_143453544 | 2.13 |
ENSRNOT00000034450
ENSRNOT00000083956 |
Krt1
Krt5
|
keratin 1 keratin 5 |
| chr5_+_127489418 | 2.03 |
ENSRNOT00000065275
|
RGD1559786
|
similar to RIKEN cDNA 0610037L13 |
| chr3_-_103345333 | 2.03 |
ENSRNOT00000071479
|
LOC100909692
|
olfactory receptor 4F3/4F16/4F29-like |
| chr3_+_11593655 | 1.98 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr4_-_82173207 | 1.94 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chrX_+_106306795 | 1.90 |
ENSRNOT00000073661
|
Gprasp1
|
G protein-coupled receptor associated sorting protein 1 |
| chr4_+_123760743 | 1.81 |
ENSRNOT00000013498
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr5_+_33784715 | 1.78 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr1_-_193700673 | 1.78 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr16_-_64745207 | 1.77 |
ENSRNOT00000032467
|
Tti2
|
TELO2 interacting protein 2 |
| chr4_+_10748745 | 1.70 |
ENSRNOT00000046142
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr10_+_57131386 | 1.67 |
ENSRNOT00000026507
|
Psmb6
|
proteasome subunit beta 6 |
| chr4_-_100213858 | 1.66 |
ENSRNOT00000015562
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr18_-_81428971 | 1.60 |
ENSRNOT00000065201
|
Zfp407
|
zinc finger protein 407 |
| chr16_-_81587076 | 1.57 |
ENSRNOT00000092470
ENSRNOT00000092721 |
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr4_+_183656013 | 1.55 |
ENSRNOT00000055442
|
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr5_-_60559533 | 1.55 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr2_+_18354542 | 1.54 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr16_+_80904125 | 1.52 |
ENSRNOT00000080832
|
Tmco3
|
transmembrane and coiled-coil domains 3 |
| chr11_+_28945575 | 1.50 |
ENSRNOT00000061600
ENSRNOT00000090364 |
AABR07033579.3
|
|
| chr8_+_67615635 | 1.49 |
ENSRNOT00000008971
|
Itga11
|
integrin subunit alpha 11 |
| chr3_+_140106766 | 1.48 |
ENSRNOT00000014046
|
Naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr3_-_103400272 | 1.48 |
ENSRNOT00000046527
|
Olr790
|
olfactory receptor 790 |
| chr7_-_15852930 | 1.45 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr7_-_11754508 | 1.44 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr2_+_196655469 | 1.38 |
ENSRNOT00000028730
|
Ctsk
|
cathepsin K |
| chr3_-_77127190 | 1.33 |
ENSRNOT00000007996
|
Olr652
|
olfactory receptor 652 |
| chr1_-_168611670 | 1.33 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr6_-_51019407 | 1.33 |
ENSRNOT00000011659
|
Gpr22
|
G protein-coupled receptor 22 |
| chr3_-_103128243 | 1.23 |
ENSRNOT00000046310
|
Olr779
|
olfactory receptor 779 |
| chr4_+_66276835 | 1.22 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr14_+_46001849 | 1.21 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr9_-_112027155 | 1.17 |
ENSRNOT00000021258
ENSRNOT00000080962 |
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr8_-_119219232 | 1.15 |
ENSRNOT00000040840
|
Tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr15_-_49005058 | 1.15 |
ENSRNOT00000019262
|
Elp3
|
elongator acetyltransferase complex subunit 3 |
| chr11_-_70211701 | 1.04 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr2_+_186630278 | 1.02 |
ENSRNOT00000021604
|
LOC365839
|
similar to elongation protein 4 homolog |
| chr15_-_37831031 | 1.01 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr11_-_28842287 | 0.97 |
ENSRNOT00000061610
|
Krtap19-5
|
keratin associated protein 19-5 |
| chr17_+_47302272 | 0.95 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr1_-_60332899 | 0.92 |
ENSRNOT00000078636
|
LOC108348087
|
vomeronasal type-1 receptor 4-like |
| chr7_-_52404774 | 0.92 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr15_-_27032498 | 0.92 |
ENSRNOT00000016702
|
Olr1609
|
olfactory receptor 1609 |
| chr13_+_52887649 | 0.92 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr8_+_115131367 | 0.91 |
ENSRNOT00000014849
|
Rpl29
|
ribosomal protein L29 |
| chr10_+_64336200 | 0.90 |
ENSRNOT00000046519
|
Rpl37
|
ribosomal protein L37 |
| chr5_+_2043583 | 0.89 |
ENSRNOT00000082813
|
Eloc
|
elongin C |
| chr2_-_189333015 | 0.84 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr2_-_189333322 | 0.84 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr6_+_54417903 | 0.81 |
ENSRNOT00000052356
|
Prps1l1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr18_-_37776453 | 0.80 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr10_+_60231027 | 0.80 |
ENSRNOT00000085543
|
Olr1482
|
olfactory receptor 1482 |
| chr8_+_99568958 | 0.78 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr4_-_162498052 | 0.76 |
ENSRNOT00000066349
|
LOC100360260
|
chromobox homolog 3-like |
| chr10_-_29196252 | 0.74 |
ENSRNOT00000083910
|
Fabp6
|
fatty acid binding protein 6 |
| chr2_+_159138758 | 0.73 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chr16_-_16762240 | 0.73 |
ENSRNOT00000080746
|
AABR07024790.1
|
|
| chr5_-_156781291 | 0.73 |
ENSRNOT00000075128
|
Cda
|
cytidine deaminase |
| chr7_-_97994586 | 0.72 |
ENSRNOT00000077616
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr4_-_82258765 | 0.72 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chr1_-_60517915 | 0.72 |
ENSRNOT00000078071
|
Vom1r11
|
vomeronasal 1 receptor 11 |
| chr8_+_45061670 | 0.70 |
ENSRNOT00000028909
|
Bsx
|
brain specific homeobox |
| chr3_-_78663607 | 0.70 |
ENSRNOT00000073134
|
Olr718
|
olfactory receptor 718 |
| chr9_-_17880706 | 0.70 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr10_-_83128297 | 0.69 |
ENSRNOT00000082160
|
Kat7
|
lysine acetyltransferase 7 |
| chr15_+_26033791 | 0.68 |
ENSRNOT00000020074
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chrX_+_111735820 | 0.68 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr1_-_149859479 | 0.68 |
ENSRNOT00000052199
|
LOC100910009
|
olfactory receptor 14A2-like |
| chr3_-_77677349 | 0.67 |
ENSRNOT00000084989
|
Olr669
|
olfactory receptor 669 |
| chr1_-_149675443 | 0.67 |
ENSRNOT00000086084
|
Olr294
|
olfactory receptor 294 |
| chr3_-_103265059 | 0.64 |
ENSRNOT00000073965
|
LOC103691873
|
putative olfactory receptor GPCRLTM7 |
| chr1_+_68239314 | 0.64 |
ENSRNOT00000070823
|
Vom1r109
|
vomeronasal 1 receptor 109 |
| chr1_-_174379566 | 0.62 |
ENSRNOT00000067388
|
Tmem9b
|
TMEM9 domain family, member B |
| chr11_+_73693814 | 0.62 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr10_-_44659707 | 0.61 |
ENSRNOT00000002064
|
RGD1559534
|
similar to Alpha enolase (2-phospho-D-glycerate hydro-lyase) |
| chr2_+_166403265 | 0.61 |
ENSRNOT00000012417
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr14_-_21898284 | 0.60 |
ENSRNOT00000036314
|
Prr27
|
proline rich 27 |
| chr1_-_149770781 | 0.56 |
ENSRNOT00000082185
|
Olr16
|
olfactory receptor 16 |
| chr6_-_92007917 | 0.56 |
ENSRNOT00000006425
ENSRNOT00000089891 |
Sos2
|
SOS Ras/Rho guanine nucleotide exchange factor 2 |
| chr14_+_812382 | 0.55 |
ENSRNOT00000041506
ENSRNOT00000039993 |
AABR07014089.1
|
|
| chrX_+_104734082 | 0.54 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr11_-_782954 | 0.52 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr8_+_48727618 | 0.51 |
ENSRNOT00000029546
|
Rps25
|
ribosomal protein s25 |
| chr3_-_76102782 | 0.51 |
ENSRNOT00000007690
|
Olr602
|
olfactory receptor 602 |
| chr3_-_73484676 | 0.51 |
ENSRNOT00000040041
|
Olr481
|
olfactory receptor 481 |
| chr1_+_65906344 | 0.50 |
ENSRNOT00000068611
|
Vom2r34
|
vomeronasal 2 receptor, 34 |
| chr7_-_3123297 | 0.49 |
ENSRNOT00000061878
|
Rab5b
|
RAB5B, member RAS oncogene family |
| chr10_+_92245442 | 0.49 |
ENSRNOT00000006808
|
Sppl2c
|
signal peptide peptidase like 2C |
| chr14_+_900696 | 0.47 |
ENSRNOT00000049544
|
Vom2r71
|
vomeronasal 2 receptor, 71 |
| chr6_-_51018050 | 0.45 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr14_-_3288017 | 0.43 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr19_-_19727081 | 0.43 |
ENSRNOT00000020700
|
Adcy7
|
adenylate cyclase 7 |
| chr14_-_44375804 | 0.42 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr7_-_119200623 | 0.41 |
ENSRNOT00000029753
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr1_-_150173316 | 0.39 |
ENSRNOT00000044676
|
Olr27
|
olfactory receptor 27 |
| chr10_-_109622745 | 0.39 |
ENSRNOT00000074354
ENSRNOT00000088317 |
Pde6g
|
phosphodiesterase 6G |
| chr10_+_63884338 | 0.36 |
ENSRNOT00000007100
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr2_+_187993758 | 0.36 |
ENSRNOT00000027182
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr16_-_62294769 | 0.35 |
ENSRNOT00000020663
|
Ppp2cb
|
protein phosphatase 2 catalytic subunit beta |
| chr14_+_4362717 | 0.35 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr1_+_172157739 | 0.34 |
ENSRNOT00000044619
|
Olr242
|
olfactory receptor 242 |
| chr8_-_36388224 | 0.34 |
ENSRNOT00000038312
|
Tirap
|
TIR domain containing adaptor protein |
| chr17_+_82430836 | 0.34 |
ENSRNOT00000083164
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr3_+_161178117 | 0.32 |
ENSRNOT00000051826
|
Spint5p
|
serine peptidase inhibitor, Kunitz type 5 |
| chr17_+_47301511 | 0.30 |
ENSRNOT00000092051
ENSRNOT00000087178 |
Nme8
|
NME/NM23 family member 8 |
| chr6_+_27535020 | 0.28 |
ENSRNOT00000076512
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr5_-_147584038 | 0.28 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr7_+_7068703 | 0.28 |
ENSRNOT00000074567
|
Olr1012
|
olfactory receptor 1012 |
| chr14_+_83889089 | 0.28 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr3_+_78876609 | 0.26 |
ENSRNOT00000049936
|
Olr726
|
olfactory receptor 726 |
| chr1_-_150262213 | 0.25 |
ENSRNOT00000049595
|
Olr32
|
olfactory receptor 32 |
| chr3_-_77170085 | 0.23 |
ENSRNOT00000042667
|
Olr654
|
olfactory receptor 654 |
| chr3_-_74749960 | 0.22 |
ENSRNOT00000046197
|
Olr491
|
olfactory receptor 491 |
| chr10_+_29606748 | 0.21 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr8_+_1459526 | 0.20 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr2_-_250862419 | 0.19 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr8_+_41987849 | 0.19 |
ENSRNOT00000073800
|
LOC100911999
|
olfactory receptor 143-like |
| chr8_+_41350182 | 0.17 |
ENSRNOT00000072823
|
LOC100911758
|
olfactory receptor 143-like |
| chr14_-_194072 | 0.17 |
ENSRNOT00000073973
|
Vmn2r112
|
vomeronasal 2, receptor 112 |
| chr14_-_89238094 | 0.17 |
ENSRNOT00000006818
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr12_+_51937175 | 0.16 |
ENSRNOT00000056780
|
Pus1
|
pseudouridylate synthase 1 |
| chr1_-_150210403 | 0.14 |
ENSRNOT00000081156
|
Olr29
|
olfactory receptor 29 |
| chr2_+_166856784 | 0.13 |
ENSRNOT00000035765
|
Otol1
|
otolin 1 |
| chr2_+_257626383 | 0.13 |
ENSRNOT00000080993
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chrX_-_79066932 | 0.13 |
ENSRNOT00000057460
|
RGD1561552
|
similar to WASP family 1 |
| chr1_-_59903339 | 0.13 |
ENSRNOT00000039037
|
Fpr-rs3
|
formyl peptide receptor, related sequence 3 |
| chr8_-_115263484 | 0.11 |
ENSRNOT00000039696
ENSRNOT00000081138 |
Iqcf3
|
IQ motif containing F3 |
| chr3_+_73235594 | 0.08 |
ENSRNOT00000082357
|
Olr463
|
olfactory receptor 463 |
| chr20_-_6556350 | 0.08 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chr3_-_78429583 | 0.08 |
ENSRNOT00000042658
|
Olr705
|
olfactory receptor 705 |
| chr1_-_168310071 | 0.08 |
ENSRNOT00000044675
|
Olr82
|
olfactory receptor 82 |
| chr8_+_82380757 | 0.08 |
ENSRNOT00000013512
|
Leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr3_-_75576520 | 0.07 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr16_+_2706428 | 0.06 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr16_+_24980723 | 0.03 |
ENSRNOT00000082142
|
Tma16
|
translation machinery associated 16 homolog |
| chr9_+_67699379 | 0.03 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr11_-_43358275 | 0.03 |
ENSRNOT00000060891
|
Olr1542
|
olfactory receptor 1542 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa11_Hoxc12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 4.3 | 25.6 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 3.1 | 9.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 2.2 | 10.9 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 1.7 | 5.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.3 | 14.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 1.3 | 5.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.2 | 3.5 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 1.1 | 17.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.9 | 2.7 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.8 | 2.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.8 | 3.9 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.8 | 6.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 4.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.6 | 3.3 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.5 | 2.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 4.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.5 | 1.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 2.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 3.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 2.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 1.5 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.3 | 5.0 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 9.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 2.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 0.7 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.2 | 1.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 13.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 2.4 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.3 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 6.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 7.2 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 13.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.6 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 2.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 1.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.6 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 1.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 2.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.1 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 7.7 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 1.5 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.6 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 2.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 4.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.1 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.8 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 2.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 14.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.3 | 10.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.8 | 25.2 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 10.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 14.3 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 4.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 2.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 15.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 21.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 5.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 2.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 6.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 16.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 3.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 10.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 25.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.1 | 9.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 2.0 | 27.7 | GO:0031432 | titin binding(GO:0031432) |
| 1.8 | 14.7 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.3 | 10.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.0 | 5.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.8 | 6.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 3.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 3.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.6 | 4.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 1.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 10.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 2.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 2.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 7.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 1.1 | GO:0038100 | nodal binding(GO:0038100) type I activin receptor binding(GO:0070698) |
| 0.4 | 1.1 | GO:0010484 | phosphorylase kinase regulator activity(GO:0008607) H3 histone acetyltransferase activity(GO:0010484) |
| 0.4 | 1.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 13.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 2.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 2.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 3.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 2.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 2.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 16.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 10.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 8.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.4 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 2.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 3.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.8 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 2.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 17.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 9.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 6.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 5.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 7.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 10.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 15.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 27.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 2.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 10.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 1.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 2.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 10.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 25.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |