Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
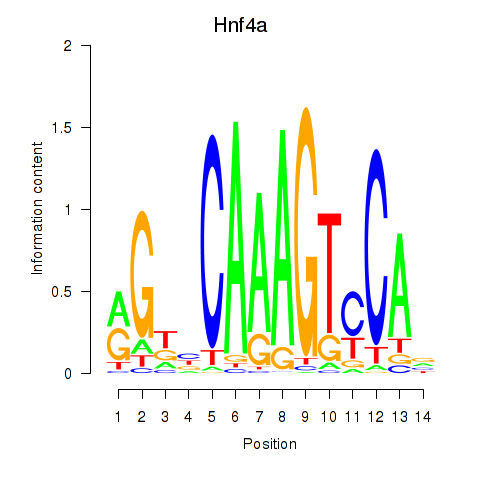
Results for Hnf4a
Z-value: 4.87
Transcription factors associated with Hnf4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf4a
|
ENSRNOG00000008895 | hepatocyte nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf4a | rn6_v1_chr3_+_159936856_159936856 | 0.94 | 4.0e-152 | Click! |
Activity profile of Hnf4a motif
Sorted Z-values of Hnf4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_60337667 | 405.58 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr1_-_80594136 | 396.22 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr7_-_123638702 | 393.99 |
ENSRNOT00000082473
ENSRNOT00000044470 ENSRNOT00000092017 |
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr4_-_161850875 | 381.34 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr10_+_89285855 | 346.00 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr1_-_259287684 | 340.66 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr13_+_56598957 | 331.69 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr1_+_100471066 | 322.07 |
ENSRNOT00000067562
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr10_+_89286047 | 321.04 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr1_+_20856187 | 314.64 |
ENSRNOT00000071726
|
Smlr1
|
small leucine-rich protein 1 |
| chr2_+_150146234 | 300.34 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr7_-_123655896 | 293.96 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr10_+_56662561 | 290.59 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr2_+_188449210 | 284.43 |
ENSRNOT00000027700
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr7_+_11490852 | 274.94 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr1_-_258766881 | 274.34 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr1_+_100470722 | 274.18 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr1_-_148119857 | 264.99 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr8_-_130550388 | 261.13 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr16_-_81822716 | 255.87 |
ENSRNOT00000026677
|
F10
|
coagulation factor X |
| chr2_+_198965685 | 249.39 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr6_+_33176778 | 245.92 |
ENSRNOT00000046811
ENSRNOT00000007371 |
Apob
|
apolipoprotein B |
| chr7_+_140781799 | 237.90 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr14_+_22142364 | 235.28 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chr10_+_71159869 | 234.27 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr1_-_198559568 | 234.10 |
ENSRNOT00000023080
|
Qprt
|
quinolinate phosphoribosyltransferase |
| chr3_-_127500709 | 231.91 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr1_+_83653234 | 231.27 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr17_-_80807181 | 226.09 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr5_+_160306727 | 221.71 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr13_+_82479998 | 219.76 |
ENSRNOT00000079872
|
F5
|
coagulation factor V |
| chrX_-_13601069 | 219.41 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr7_-_3386522 | 218.05 |
ENSRNOT00000010760
|
Mettl7b
|
methyltransferase like 7B |
| chr5_+_137357674 | 216.81 |
ENSRNOT00000092813
|
RGD1305347
|
similar to RIKEN cDNA 2610528J11 |
| chr6_+_128050250 | 215.35 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr13_+_75175254 | 209.52 |
ENSRNOT00000044008
|
Sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr12_+_47407811 | 206.09 |
ENSRNOT00000001565
|
Hnf1a
|
HNF1 homeobox A |
| chr19_+_37052556 | 202.76 |
ENSRNOT00000019072
|
Ces2h
|
carboxylesterase 2H |
| chr3_-_101465995 | 199.82 |
ENSRNOT00000080175
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr11_+_65022100 | 192.91 |
ENSRNOT00000003934
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr10_+_64952119 | 192.72 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr17_-_9762813 | 191.51 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr3_-_5802129 | 184.25 |
ENSRNOT00000009555
|
Sardh
|
sarcosine dehydrogenase |
| chr13_+_93684437 | 182.36 |
ENSRNOT00000005005
|
Kmo
|
kynurenine 3-monooxygenase |
| chr10_-_7200499 | 178.08 |
ENSRNOT00000003633
|
Abat
|
4-aminobutyrate aminotransferase |
| chr10_-_31790263 | 177.79 |
ENSRNOT00000059468
|
Timd2
|
T-cell immunoglobulin and mucin domain containing 2 |
| chr1_+_264741911 | 176.50 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr1_-_224389389 | 175.38 |
ENSRNOT00000077408
ENSRNOT00000050010 |
UST4r
|
integral membrane transport protein UST4r |
| chr9_+_20048121 | 170.70 |
ENSRNOT00000014791
|
Mep1a
|
meprin 1 subunit alpha |
| chr8_-_103608913 | 170.22 |
ENSRNOT00000013209
|
Pls1
|
plastin 1 |
| chr1_+_88955440 | 165.27 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_-_258877045 | 164.94 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr10_-_15459897 | 164.80 |
ENSRNOT00000027518
|
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr15_-_34444244 | 163.29 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr11_+_74057361 | 162.19 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr14_+_7113544 | 160.98 |
ENSRNOT00000038188
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_88955135 | 157.54 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr2_+_227080924 | 156.96 |
ENSRNOT00000029871
|
Fabp2
|
fatty acid binding protein 2 |
| chr18_-_24823837 | 151.46 |
ENSRNOT00000021405
ENSRNOT00000090390 |
Myo7b
|
myosin VIIb |
| chr4_+_100218661 | 140.26 |
ENSRNOT00000079415
|
Tmem150a
|
transmembrane protein 150A |
| chr14_+_87448692 | 133.67 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr20_-_4542073 | 128.45 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr2_+_93669765 | 127.46 |
ENSRNOT00000045438
|
Slc10a5
|
solute carrier family 10, member 5 |
| chr8_-_119012671 | 124.03 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr1_-_224698514 | 121.14 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr4_-_176679815 | 118.75 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr18_-_15089988 | 116.25 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr11_+_87058616 | 115.31 |
ENSRNOT00000002576
ENSRNOT00000082855 |
Prodh1
|
proline dehydrogenase 1 |
| chr10_+_65767053 | 115.01 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chr2_-_225389120 | 111.18 |
ENSRNOT00000016739
|
Abcd3
|
ATP binding cassette subfamily D member 3 |
| chr1_-_207811008 | 109.74 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr20_-_4508197 | 108.86 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr6_+_56846789 | 107.66 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr20_-_2678141 | 106.29 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr2_+_53109684 | 105.00 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chr2_+_188449718 | 103.69 |
ENSRNOT00000065791
|
Pklr
|
pyruvate kinase, liver and RBC |
| chr6_+_8284878 | 101.77 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr1_+_14224393 | 101.64 |
ENSRNOT00000016037
|
Perp
|
PERP, TP53 apoptosis effector |
| chr7_+_143754892 | 96.43 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr5_+_157759416 | 91.82 |
ENSRNOT00000024063
ENSRNOT00000083629 |
Akr7a2
|
aldo-keto reductase family 7, member A2 |
| chr1_-_82409639 | 90.80 |
ENSRNOT00000031326
|
Erich4
|
glutamate-rich 4 |
| chr3_-_64024205 | 90.19 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_+_242572533 | 89.79 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr14_+_77079402 | 89.19 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr8_-_119265157 | 88.78 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr11_+_87204175 | 86.33 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr5_+_167141875 | 85.83 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr16_+_20555395 | 80.14 |
ENSRNOT00000026652
|
Gdf15
|
growth differentiation factor 15 |
| chr13_+_110677810 | 71.37 |
ENSRNOT00000006340
|
Slc30a1
|
solute carrier family 30 member 1 |
| chr13_+_84334598 | 70.25 |
ENSRNOT00000004986
|
Gpa33
|
glycoprotein A33 |
| chr2_+_60180215 | 67.92 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr13_+_89586283 | 67.85 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr13_-_53870428 | 67.75 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrX_+_54266687 | 67.37 |
ENSRNOT00000058309
|
LOC100363193
|
LRRGT00076-like |
| chr4_-_22192474 | 65.71 |
ENSRNOT00000043822
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr4_+_62380914 | 64.55 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr5_-_137112927 | 64.30 |
ENSRNOT00000078302
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr6_+_26051396 | 61.83 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr12_-_17972737 | 60.98 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr3_-_5481144 | 60.40 |
ENSRNOT00000078429
|
Surf4
|
surfeit 4 |
| chr5_+_167142182 | 58.39 |
ENSRNOT00000024054
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr16_+_26859397 | 58.01 |
ENSRNOT00000044171
|
Msmo1
|
methylsterol monooxygenase 1 |
| chr5_-_131860637 | 56.80 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_+_142950540 | 56.60 |
ENSRNOT00000025621
|
Slc28a1
|
solute carrier family 28 member 1 |
| chr8_-_48582353 | 54.88 |
ENSRNOT00000011582
|
Pdzd3
|
PDZ domain containing 3 |
| chr4_+_88832178 | 54.67 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr10_+_61432819 | 53.09 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr5_-_101138427 | 50.59 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr3_+_44806106 | 48.55 |
ENSRNOT00000035158
|
Upp2
|
uridine phosphorylase 2 |
| chr7_+_119626637 | 47.91 |
ENSRNOT00000000201
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr20_-_4316715 | 47.28 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr2_-_154542557 | 47.18 |
ENSRNOT00000013392
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr4_+_49056010 | 43.74 |
ENSRNOT00000038566
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_47281421 | 43.37 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr12_+_2134022 | 42.55 |
ENSRNOT00000001305
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr3_+_81287242 | 41.28 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr20_-_4368693 | 40.71 |
ENSRNOT00000000501
|
Rnf5
|
ring finger protein 5 |
| chr15_+_33121273 | 40.59 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr19_+_26106838 | 40.56 |
ENSRNOT00000035987
|
Hook2
|
hook microtubule-tethering protein 2 |
| chr6_-_103313074 | 38.79 |
ENSRNOT00000083677
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr2_+_58462588 | 38.74 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr8_+_23014956 | 37.00 |
ENSRNOT00000018009
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr13_-_53108713 | 36.98 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr2_-_154542919 | 36.75 |
ENSRNOT00000076880
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr7_-_120077612 | 35.52 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr4_+_85551502 | 35.35 |
ENSRNOT00000087191
ENSRNOT00000015692 |
Aqp1
|
aquaporin 1 |
| chr4_-_155401480 | 33.36 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr6_-_79306443 | 32.62 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr10_+_62981297 | 32.33 |
ENSRNOT00000031618
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr12_+_47254484 | 31.93 |
ENSRNOT00000001556
|
Acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr6_-_44363915 | 31.48 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr3_-_161272460 | 29.34 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr1_+_222311253 | 28.16 |
ENSRNOT00000028749
|
Macrod1
|
MACRO domain containing 1 |
| chr1_-_164659992 | 26.22 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr2_+_153803349 | 25.51 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chr13_+_50434886 | 25.38 |
ENSRNOT00000076857
|
Sox13
|
SRY box 13 |
| chr1_-_127599257 | 25.26 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr8_+_79489790 | 25.21 |
ENSRNOT00000091722
|
Prtg
|
protogenin |
| chr1_+_258210344 | 25.10 |
ENSRNOT00000001990
|
LOC100361492
|
cytochrome P450, family 2, subfamily c, polypeptide 55-like |
| chr2_-_219741886 | 24.67 |
ENSRNOT00000085122
|
Slc35a3
|
solute carrier family 35 member A3 |
| chr8_-_85840818 | 23.69 |
ENSRNOT00000013608
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_69201380 | 23.09 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr15_-_46432965 | 19.60 |
ENSRNOT00000014320
|
Gata4
|
GATA binding protein 4 |
| chr8_+_70112925 | 19.53 |
ENSRNOT00000082401
|
Megf11
|
multiple EGF-like-domains 11 |
| chr6_+_22167919 | 18.93 |
ENSRNOT00000007655
|
Nlrc4
|
NLR family, CARD domain containing 4 |
| chr10_-_110701137 | 18.67 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr1_-_175420106 | 17.77 |
ENSRNOT00000013126
ENSRNOT00000077125 |
Sbf2
|
SET binding factor 2 |
| chr12_+_41600005 | 17.42 |
ENSRNOT00000001872
|
Plbd2
|
phospholipase B domain containing 2 |
| chr7_-_14054639 | 16.98 |
ENSRNOT00000039852
|
Ilvbl
|
ilvB acetolactate synthase like |
| chr13_+_50434702 | 16.30 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr3_-_45210474 | 16.13 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr3_-_120106697 | 15.29 |
ENSRNOT00000020354
|
Prom2
|
prominin 2 |
| chr8_-_49271834 | 14.91 |
ENSRNOT00000085022
|
Ube4a
|
ubiquitination factor E4A |
| chr8_+_22035256 | 14.70 |
ENSRNOT00000028066
|
Icam1
|
intercellular adhesion molecule 1 |
| chr8_+_72743426 | 14.51 |
ENSRNOT00000072573
|
Rps27l
|
ribosomal protein S27-like |
| chr1_-_219745654 | 14.37 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr5_+_9230984 | 14.25 |
ENSRNOT00000009136
|
Vcpip1
|
valosin containing protein interacting protein 1 |
| chr9_-_81565416 | 13.53 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr5_+_137257637 | 12.84 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr10_+_34519790 | 12.11 |
ENSRNOT00000052360
|
Mgat1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr3_+_95133713 | 11.88 |
ENSRNOT00000067940
|
Wt1
|
Wilms tumor 1 |
| chr7_-_70842405 | 9.77 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr10_-_15381691 | 9.08 |
ENSRNOT00000027439
|
Rab11fip3
|
RAB11 family interacting protein 3 |
| chr7_-_142062870 | 9.00 |
ENSRNOT00000026531
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr11_-_70833577 | 8.96 |
ENSRNOT00000002428
|
Osbpl11
|
oxysterol binding protein-like 11 |
| chr3_-_146812989 | 8.51 |
ENSRNOT00000011315
|
Nanp
|
N-acetylneuraminic acid phosphatase |
| chr10_-_63274640 | 7.89 |
ENSRNOT00000005129
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_257376756 | 7.46 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr4_+_157181795 | 7.30 |
ENSRNOT00000017090
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr10_-_56185857 | 6.26 |
ENSRNOT00000014261
|
Wrap53
|
WD repeat containing, antisense to TP53 |
| chr2_-_232178533 | 5.04 |
ENSRNOT00000082173
ENSRNOT00000055607 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_210873024 | 4.28 |
ENSRNOT00000026051
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chrX_+_33884499 | 4.10 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr15_-_12513931 | 4.04 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr13_+_48790509 | 3.81 |
ENSRNOT00000065094
|
Elk4
|
ELK4, ETS transcription factor |
| chr3_+_154507035 | 3.13 |
ENSRNOT00000017265
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr10_+_89635675 | 2.89 |
ENSRNOT00000028179
|
RGD1561590
|
similar to SAP18 |
| chr2_-_80293181 | 2.58 |
ENSRNOT00000016111
|
Otulin
|
OTU deubiquitinase with linear linkage specificity |
| chr5_+_150032999 | 2.57 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr20_+_4162055 | 2.45 |
ENSRNOT00000081236
|
Btnl3
|
butyrophilin-like 3 |
| chrX_-_157172068 | 2.32 |
ENSRNOT00000087962
|
Dusp9
|
dual specificity phosphatase 9 |
| chr14_-_84937725 | 1.66 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr13_+_48790767 | 1.15 |
ENSRNOT00000087504
|
Elk4
|
ELK4, ETS transcription factor |
| chr9_+_76440186 | 1.00 |
ENSRNOT00000049479
|
RGD1562431
|
similar to chromosome 20 open reading frame 81 |
| chr5_+_74727494 | 0.51 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr10_-_89918427 | 0.33 |
ENSRNOT00000084217
|
Dusp3
|
dual specificity phosphatase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 198.7 | 596.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 146.0 | 438.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 135.2 | 405.6 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 122.6 | 367.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 110.1 | 440.4 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 84.2 | 252.5 | GO:0097187 | dentinogenesis(GO:0097187) |
| 69.4 | 416.5 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 66.7 | 667.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 62.3 | 249.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 58.0 | 231.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 53.2 | 1276.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 48.9 | 586.9 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 48.5 | 388.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 48.2 | 192.7 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 45.2 | 226.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 44.3 | 221.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 43.9 | 219.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 42.6 | 170.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 36.9 | 184.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 35.6 | 178.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 35.1 | 245.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 33.6 | 235.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 33.4 | 300.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 33.3 | 199.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 31.9 | 31.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 31.9 | 351.2 | GO:0015747 | urate transport(GO:0015747) |
| 30.3 | 151.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 28.8 | 144.2 | GO:0071332 | fructose transport(GO:0015755) cellular response to fructose stimulus(GO:0071332) |
| 24.4 | 219.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 23.8 | 71.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 23.3 | 209.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 21.6 | 86.3 | GO:0015746 | citrate transport(GO:0015746) |
| 20.6 | 61.8 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 18.3 | 274.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 18.3 | 54.9 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 16.4 | 65.7 | GO:1990962 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 16.0 | 47.9 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 15.0 | 164.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 14.1 | 56.6 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 13.9 | 111.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 13.5 | 67.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 12.9 | 38.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 12.1 | 96.4 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 11.8 | 35.4 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 11.7 | 128.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 11.5 | 91.8 | GO:0030647 | aminoglycoside antibiotic metabolic process(GO:0030647) |
| 11.1 | 33.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 10.7 | 64.3 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 10.5 | 31.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 10.5 | 83.9 | GO:0051182 | coenzyme transport(GO:0051182) |
| 10.5 | 115.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 9.2 | 101.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 9.0 | 107.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 8.9 | 124.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 8.5 | 42.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 8.2 | 294.0 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 8.1 | 89.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 8.1 | 48.5 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 8.0 | 215.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 7.9 | 47.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 7.7 | 38.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 7.7 | 23.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 7.5 | 67.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 7.0 | 28.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 7.0 | 153.6 | GO:0097286 | iron ion import(GO:0097286) |
| 6.6 | 105.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 6.6 | 26.2 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 6.5 | 19.6 | GO:1905204 | septum secundum development(GO:0003285) embryonic heart tube anterior/posterior pattern specification(GO:0035054) negative regulation of connective tissue replacement(GO:1905204) |
| 6.4 | 25.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 6.3 | 6.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 6.2 | 24.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 6.1 | 381.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 5.8 | 127.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 5.3 | 137.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 4.9 | 29.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 4.6 | 23.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 4.4 | 88.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 4.4 | 127.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 4.3 | 60.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 4.1 | 40.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 4.0 | 12.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 4.0 | 80.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 4.0 | 11.9 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 3.8 | 41.7 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 3.7 | 14.7 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 3.3 | 53.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 3.2 | 140.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 3.1 | 93.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 3.1 | 15.3 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 2.9 | 14.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 2.8 | 8.5 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 2.8 | 90.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 2.8 | 532.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 2.7 | 18.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 2.6 | 116.2 | GO:1901998 | toxin transport(GO:1901998) |
| 2.4 | 161.0 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 2.3 | 118.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 2.3 | 56.8 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.9 | 43.4 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 1.8 | 9.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 1.8 | 37.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.6 | 58.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 1.5 | 9.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.3 | 40.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 1.2 | 23.7 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 1.2 | 286.4 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.9 | 32.6 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.9 | 2.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.7 | 19.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.7 | 5.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 4.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 3.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.6 | 198.2 | GO:0032259 | methylation(GO:0032259) |
| 0.6 | 92.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.5 | 14.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 14.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 7.3 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.2 | 13.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 2.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 86.1 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.1 | 2.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 10.2 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 107.0 | 642.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 24.3 | 170.2 | GO:1990357 | terminal web(GO:1990357) |
| 19.2 | 115.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 15.7 | 157.0 | GO:0045179 | apical cortex(GO:0045179) |
| 15.6 | 217.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 10.2 | 264.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 9.7 | 48.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 9.2 | 219.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 8.8 | 35.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 8.1 | 40.6 | GO:0070695 | FHF complex(GO:0070695) |
| 7.4 | 140.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 7.4 | 206.1 | GO:0045120 | pronucleus(GO:0045120) |
| 7.1 | 542.3 | GO:0005811 | lipid particle(GO:0005811) |
| 6.1 | 42.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 5.1 | 40.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 5.1 | 65.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 5.1 | 667.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 4.3 | 2885.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 4.2 | 557.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 3.8 | 18.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 3.6 | 268.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 3.2 | 231.8 | GO:0005903 | brush border(GO:0005903) |
| 3.2 | 47.3 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 3.1 | 999.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 2.7 | 290.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 2.2 | 142.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 1.7 | 598.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 1.5 | 43.4 | GO:0008305 | integrin complex(GO:0008305) |
| 1.3 | 60.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 1.2 | 313.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.9 | 50.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.6 | 63.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.6 | 100.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.6 | 1376.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.5 | 98.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.5 | 2.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 13.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 346.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.4 | 14.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 130.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.4 | 87.6 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.3 | 6.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.3 | 1278.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.2 | 72.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 9.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 26.8 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 146.0 | 438.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 133.4 | 667.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 132.1 | 396.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 97.0 | 388.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 81.1 | 405.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 76.3 | 381.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 58.5 | 234.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 58.0 | 231.9 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 49.2 | 245.9 | GO:0035473 | lipase binding(GO:0035473) |
| 48.2 | 192.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 44.7 | 268.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 42.5 | 1276.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 41.2 | 164.8 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 38.6 | 385.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 38.3 | 191.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 33.6 | 235.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 32.3 | 226.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 25.8 | 283.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 25.6 | 639.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 22.6 | 67.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 21.6 | 86.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 21.2 | 127.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 20.6 | 596.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 20.3 | 182.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 18.9 | 56.6 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 18.7 | 261.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 18.4 | 91.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 17.8 | 88.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 17.2 | 274.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 17.1 | 221.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 17.0 | 118.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 16.8 | 83.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 16.0 | 144.2 | GO:0070061 | fructose binding(GO:0070061) |
| 16.0 | 47.9 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 15.4 | 107.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 15.0 | 120.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 15.0 | 300.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 15.0 | 105.0 | GO:0008430 | selenium binding(GO:0008430) |
| 14.9 | 133.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 11.5 | 184.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 8.7 | 43.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 8.1 | 178.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 7.9 | 47.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 7.1 | 35.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 6.9 | 54.9 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 6.8 | 520.3 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 6.5 | 71.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 6.3 | 25.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 6.2 | 67.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 5.8 | 23.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 5.7 | 56.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 5.6 | 33.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 5.0 | 49.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 4.6 | 128.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 4.3 | 260.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 4.1 | 32.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 4.1 | 40.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 4.0 | 206.1 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 4.0 | 199.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 3.9 | 42.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 3.7 | 29.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 3.3 | 26.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 3.3 | 19.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 3.0 | 38.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 3.0 | 11.9 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 2.8 | 17.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 2.4 | 292.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 2.3 | 139.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 2.3 | 124.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 2.1 | 61.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 1.8 | 384.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 1.7 | 58.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 1.6 | 14.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.6 | 237.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 1.5 | 24.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 1.5 | 14.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 1.5 | 38.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 1.5 | 202.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 1.1 | 23.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.1 | 151.5 | GO:0003774 | motor activity(GO:0003774) |
| 1.1 | 4.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.0 | 102.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 1.0 | 198.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 1.0 | 58.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.8 | 5.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.8 | 18.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.7 | 148.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.6 | 28.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.4 | 15.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.4 | 17.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 17.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.3 | 2.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 51.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 6.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 16.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 41.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 14.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 27.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 40.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 5.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 46.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 43.5 | GO:0016491 | oxidoreductase activity(GO:0016491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.6 | 1629.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 8.7 | 270.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 7.7 | 115.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 7.1 | 128.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 6.0 | 245.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 5.2 | 1203.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 2.9 | 43.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 2.5 | 153.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 1.6 | 67.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 1.3 | 86.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 1.1 | 171.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.9 | 67.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.6 | 38.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.5 | 23.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 19.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 33.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 11.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 6.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 50.5 | 807.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 37.8 | 642.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 34.9 | 454.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 26.4 | 343.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 23.0 | 896.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 21.8 | 261.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 20.3 | 182.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 18.8 | 226.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 18.1 | 235.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 16.3 | 636.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 15.6 | 233.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 12.3 | 221.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 6.7 | 133.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 6.1 | 54.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 5.9 | 152.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 5.8 | 93.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 5.6 | 191.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 4.9 | 29.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 4.3 | 234.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 3.7 | 37.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 3.6 | 651.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 3.2 | 67.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 3.1 | 214.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 2.9 | 35.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 2.8 | 62.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 2.6 | 283.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 2.5 | 58.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 2.5 | 66.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 2.3 | 31.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 2.0 | 38.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 1.8 | 19.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.6 | 118.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 1.6 | 129.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 1.0 | 18.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.8 | 80.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.7 | 13.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 7.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 4.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 6.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 12.6 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 3.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |