Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
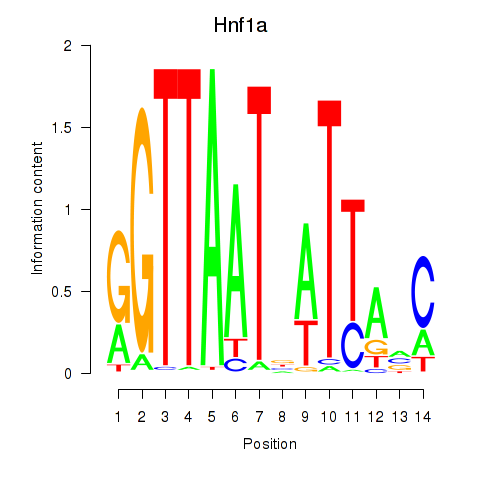
Results for Hnf1a
Z-value: 5.04
Transcription factors associated with Hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1a
|
ENSRNOG00000001183 | HNF1 homeobox A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1a | rn6_v1_chr12_+_47407811_47407811 | 0.84 | 1.4e-85 | Click! |
Activity profile of Hnf1a motif
Sorted Z-values of Hnf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_19132208 | 885.57 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr14_+_20266891 | 500.56 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr1_+_213511874 | 488.37 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr20_-_5123073 | 472.26 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr2_+_182006242 | 467.11 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr6_-_127620296 | 443.67 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr10_+_89285855 | 437.75 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr3_+_159902441 | 419.40 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr18_-_38185812 | 415.85 |
ENSRNOT00000017969
|
Spink1l
|
serine peptidase inhibitor, Kazal type 1-like |
| chr3_-_14229067 | 410.25 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr10_+_89286047 | 407.23 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr1_+_261291870 | 393.37 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr3_-_80543031 | 390.28 |
ENSRNOT00000022233
|
F2
|
coagulation factor II |
| chrX_+_71272042 | 389.02 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr14_-_19191863 | 386.02 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr1_-_48563776 | 381.91 |
ENSRNOT00000023368
|
Plg
|
plasminogen |
| chr2_-_182035032 | 377.55 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr14_+_87448692 | 363.25 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr6_-_127534247 | 361.06 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr13_+_91080341 | 355.95 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr2_+_235264219 | 344.36 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr14_-_80973456 | 335.31 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr13_+_56598957 | 333.52 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr13_-_91427575 | 323.72 |
ENSRNOT00000012092
|
Apcs
|
amyloid P component, serum |
| chr9_+_95295701 | 300.42 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_277068761 | 299.72 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr8_-_77398156 | 292.65 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr6_-_127508452 | 288.21 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr1_+_88955440 | 273.76 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr1_+_88955135 | 262.20 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr3_+_159936856 | 258.71 |
ENSRNOT00000078703
|
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr20_-_27117663 | 258.28 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr13_-_50549981 | 252.95 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr1_+_250426158 | 252.79 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr13_-_47377703 | 251.28 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chr6_-_26385761 | 250.30 |
ENSRNOT00000073228
|
Gckr
|
glucokinase regulator |
| chr15_+_28023018 | 247.86 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
| chr2_-_100372252 | 213.96 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_141579871 | 207.38 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr5_-_79008363 | 193.35 |
ENSRNOT00000010040
|
Kif12
|
kinesin family member 12 |
| chrX_-_32153794 | 192.65 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr4_+_68849033 | 189.16 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr1_+_107262659 | 181.29 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr14_+_22251499 | 180.79 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr4_+_175814118 | 178.51 |
ENSRNOT00000013409
ENSRNOT00000013514 |
Slco1b2
|
solute carrier organic anion transporter family, member 1B2 |
| chr14_-_19159923 | 177.56 |
ENSRNOT00000003879
|
Afp
|
alpha-fetoprotein |
| chr17_-_69711689 | 169.21 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr5_+_33784715 | 166.86 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr3_-_127500709 | 157.33 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr2_-_88763733 | 152.94 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr1_+_214203861 | 143.78 |
ENSRNOT00000023219
|
Rassf7
|
Ras association domain family member 7 |
| chr8_-_84320714 | 138.81 |
ENSRNOT00000079356
ENSRNOT00000088487 |
Tinag
|
tubulointerstitial nephritis antigen |
| chr19_+_568287 | 131.24 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr7_-_68549763 | 122.19 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr14_-_82975263 | 117.95 |
ENSRNOT00000024165
|
Slc5a1
|
solute carrier family 5 member 1 |
| chr4_-_51199570 | 109.24 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr20_+_41266566 | 106.88 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr8_+_22856539 | 105.62 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr13_+_42008842 | 103.82 |
ENSRNOT00000038811
|
Gpr39
|
G protein-coupled receptor 39 |
| chr17_-_10004321 | 100.08 |
ENSRNOT00000042394
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr17_+_69634890 | 97.96 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr2_-_166682325 | 97.39 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr11_-_80981415 | 88.85 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr10_-_71491743 | 88.65 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr1_-_266428239 | 88.23 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_-_88660449 | 80.53 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_+_117421604 | 76.41 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr2_-_158156444 | 73.77 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_-_158156150 | 72.43 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr20_+_32717564 | 69.96 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr8_+_49713190 | 68.40 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr4_-_82295470 | 64.30 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr4_-_82209933 | 62.66 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr3_-_11452529 | 59.88 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr19_-_54761670 | 59.72 |
ENSRNOT00000025848
|
Ca5a
|
carbonic anhydrase 5A |
| chr13_-_53870428 | 51.83 |
ENSRNOT00000000812
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr5_+_157416897 | 50.70 |
ENSRNOT00000023358
|
Rnf186
|
ring finger protein 186 |
| chr19_-_39267928 | 48.78 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_88553086 | 46.95 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_+_56887987 | 38.28 |
ENSRNOT00000041288
|
Gdnf
|
glial cell derived neurotrophic factor |
| chr5_-_158439078 | 33.30 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr7_-_98270110 | 32.71 |
ENSRNOT00000064847
|
Anxa13
|
annexin A13 |
| chr6_+_101603319 | 25.32 |
ENSRNOT00000030470
|
Gphn
|
gephyrin |
| chr15_-_52320385 | 24.94 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr8_-_127912860 | 12.05 |
ENSRNOT00000040498
|
LOC685081
|
similar to solute carrier family 22 (organic cation transporter), member 13 |
| chr9_+_119542328 | 7.57 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr11_+_64882288 | 6.10 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 178.7 | 536.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 169.5 | 678.1 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 168.9 | 844.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 157.4 | 472.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 147.9 | 443.7 | GO:0033986 | response to methanol(GO:0033986) |
| 130.1 | 390.3 | GO:1905225 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) response to thyrotropin-releasing hormone(GO:1905225) |
| 127.3 | 381.9 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 127.0 | 380.9 | GO:0010034 | response to acetate(GO:0010034) |
| 98.3 | 393.4 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 97.7 | 488.4 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 86.1 | 258.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 84.5 | 845.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 83.8 | 251.3 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 80.9 | 323.7 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 74.9 | 299.7 | GO:1904975 | response to bleomycin(GO:1904975) |
| 68.4 | 410.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 64.3 | 386.0 | GO:0070541 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) response to platinum ion(GO:0070541) |
| 63.2 | 252.8 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 59.3 | 355.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 50.1 | 250.3 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) negative regulation of glucokinase activity(GO:0033132) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) negative regulation of hexokinase activity(GO:1903300) |
| 39.3 | 157.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 37.0 | 481.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 34.6 | 103.8 | GO:0035483 | gastric emptying(GO:0035483) |
| 33.8 | 1386.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 29.9 | 448.9 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 29.6 | 88.8 | GO:1990743 | protein sialylation(GO:1990743) |
| 26.0 | 415.9 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 25.5 | 76.4 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 25.0 | 100.1 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 24.4 | 122.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 24.3 | 97.4 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 23.6 | 117.9 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 21.4 | 192.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 17.3 | 207.4 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 14.4 | 361.1 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 14.0 | 363.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 13.2 | 344.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 12.8 | 140.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 12.7 | 177.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 11.5 | 447.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 10.4 | 51.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 9.6 | 38.3 | GO:0072107 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 8.4 | 25.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 7.3 | 131.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 7.0 | 105.6 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 7.0 | 70.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 6.8 | 109.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 6.2 | 24.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 4.3 | 68.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 2.3 | 453.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 1.9 | 64.3 | GO:0060065 | uterus development(GO:0060065) |
| 1.7 | 247.9 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 1.6 | 59.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 1.5 | 106.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 1.0 | 214.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.9 | 12.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.8 | 193.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.7 | 143.8 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.2 | 7.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 60.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.2 | 179.5 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.1 | 123.9 | GO:0016192 | vesicle-mediated transport(GO:0016192) |
| 0.0 | 6.1 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 105.6 | 844.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 95.5 | 381.9 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 94.5 | 472.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 68.4 | 410.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 50.3 | 251.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 36.1 | 252.8 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 21.1 | 2486.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 18.5 | 389.0 | GO:0005922 | connexon complex(GO:0005922) |
| 17.2 | 292.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 16.0 | 207.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 12.2 | 97.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 9.9 | 88.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 9.3 | 1271.3 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 8.3 | 24.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 5.7 | 68.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 3.1 | 355.9 | GO:0030175 | filopodium(GO:0030175) |
| 3.1 | 422.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 3.0 | 335.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 2.8 | 193.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 2.0 | 794.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 2.0 | 170.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 1.7 | 2318.4 | GO:0005615 | extracellular space(GO:0005615) |
| 1.3 | 595.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 1.1 | 157.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 1.0 | 132.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.6 | 791.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.4 | 458.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.4 | 33.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 666.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.2 | 97.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.2 | 120.8 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 634.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 67.2 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.1 | 7.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 178.7 | 536.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 169.0 | 845.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 136.7 | 410.2 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 98.3 | 393.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 97.1 | 679.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 83.4 | 500.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 56.5 | 678.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 49.8 | 298.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 48.8 | 390.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 48.2 | 192.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 47.3 | 189.2 | GO:0016160 | amylase activity(GO:0016160) |
| 40.4 | 363.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 39.3 | 157.3 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 35.8 | 250.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 30.5 | 488.4 | GO:0010181 | FMN binding(GO:0010181) |
| 29.4 | 381.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 27.8 | 389.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 25.5 | 76.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 24.4 | 122.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 23.5 | 140.8 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 22.7 | 386.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 22.2 | 88.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 20.9 | 251.3 | GO:0001848 | complement binding(GO:0001848) |
| 19.5 | 97.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 15.5 | 481.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 13.0 | 1508.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 12.5 | 100.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 10.1 | 885.6 | GO:0019842 | vitamin binding(GO:0019842) |
| 9.0 | 207.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 7.6 | 405.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 6.8 | 109.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 6.7 | 59.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 6.5 | 870.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 6.3 | 88.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 5.9 | 447.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 5.2 | 472.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 4.9 | 68.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 4.6 | 59.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 4.1 | 32.7 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 3.8 | 265.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 3.0 | 635.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 3.0 | 252.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 2.3 | 247.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 2.2 | 193.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.9 | 106.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 1.7 | 38.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 1.4 | 236.6 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.9 | 12.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.9 | 24.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.8 | 181.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.7 | 105.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 134.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.3 | 201.2 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.2 | 131.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 7.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 50.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 51.6 | 2840.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 27.2 | 844.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 9.3 | 381.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 8.9 | 443.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 7.0 | 355.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 6.8 | 202.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 4.0 | 916.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 3.0 | 181.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 2.6 | 64.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 2.4 | 207.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 1.9 | 100.1 | PID FGF PATHWAY | FGF signaling pathway |
| 1.5 | 38.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 1.1 | 335.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.7 | 138.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 162.8 | 488.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 98.0 | 1568.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 45.8 | 595.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 38.6 | 386.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 37.3 | 745.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 36.5 | 766.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 33.0 | 892.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 25.9 | 155.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 25.3 | 481.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 24.3 | 1095.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 21.6 | 389.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 17.8 | 588.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 17.2 | 292.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 16.5 | 231.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 8.1 | 323.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 7.1 | 100.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 6.7 | 181.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 5.9 | 88.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 5.0 | 443.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 4.3 | 51.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 1.7 | 252.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 1.4 | 68.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.3 | 117.9 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.8 | 157.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.7 | 76.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.5 | 7.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.5 | 25.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 63.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |