Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
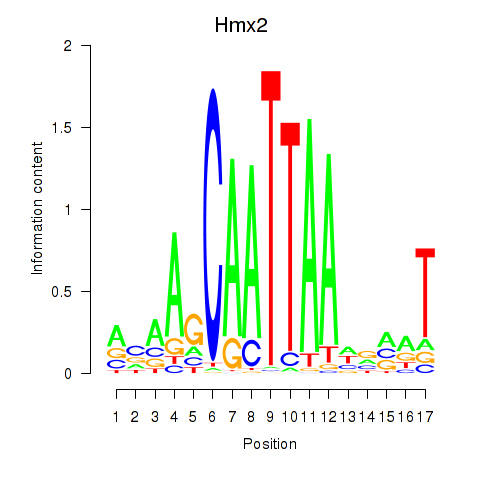
Results for Hmx2
Z-value: 0.50
Transcription factors associated with Hmx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx2
|
ENSRNOG00000020638 | H6 family homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx2 | rn6_v1_chr1_+_203515609_203515609 | 0.43 | 1.6e-15 | Click! |
Activity profile of Hmx2 motif
Sorted Z-values of Hmx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_61814974 | 17.20 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr17_-_9762813 | 16.45 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr3_-_937102 | 13.33 |
ENSRNOT00000007471
|
Hnmt
|
histamine N-methyltransferase |
| chr4_+_22859622 | 11.72 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr2_-_231648122 | 10.50 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr17_-_69404323 | 10.33 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr7_+_71709157 | 10.07 |
ENSRNOT00000008211
|
Cpq
|
carboxypeptidase Q |
| chr4_-_171591882 | 10.03 |
ENSRNOT00000009328
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr20_-_27117663 | 9.98 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr9_-_105693357 | 9.75 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chr8_+_91464229 | 9.26 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr13_+_48575091 | 8.61 |
ENSRNOT00000009838
|
Pm20d1
|
peptidase M20 domain containing 1 |
| chr3_-_120011364 | 7.80 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chrX_+_110789269 | 7.62 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_66898946 | 7.59 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr14_+_37130743 | 7.31 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr19_-_37440471 | 7.25 |
ENSRNOT00000066636
|
Zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr3_-_14112851 | 7.00 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr3_+_159368273 | 6.97 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_-_727691 | 6.91 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr5_-_17061837 | 6.68 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr18_-_61057365 | 6.56 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr5_-_113621134 | 6.37 |
ENSRNOT00000037206
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr7_+_121408829 | 6.24 |
ENSRNOT00000023434
|
Mgat3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr6_-_122897997 | 6.01 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr13_-_111972603 | 5.97 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr18_-_37132223 | 5.83 |
ENSRNOT00000091634
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_117454769 | 5.71 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr6_+_72891725 | 5.65 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr10_-_10767389 | 5.42 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr5_+_154650632 | 5.39 |
ENSRNOT00000072132
|
Zfp46
|
zinc finger protein 46 |
| chr3_+_140024043 | 5.26 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_+_217173199 | 5.26 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr5_-_17061361 | 4.99 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr6_+_73553210 | 4.96 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr8_+_100260049 | 4.87 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr4_+_88694583 | 4.84 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr9_+_47536824 | 4.73 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr10_+_49231730 | 4.54 |
ENSRNOT00000065335
|
Trim16
|
tripartite motif-containing 16 |
| chr1_-_126211439 | 4.50 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr8_+_118066988 | 4.45 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr11_+_24266345 | 4.43 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr19_-_56983807 | 4.41 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chr3_-_153042395 | 4.39 |
ENSRNOT00000055232
|
Ndrg3
|
NDRG family member 3 |
| chr1_-_52495582 | 4.31 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chrX_+_5825711 | 4.30 |
ENSRNOT00000037360
|
Efhc2
|
EF-hand domain containing 2 |
| chrM_+_9451 | 4.28 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrX_-_124516705 | 4.25 |
ENSRNOT00000061493
|
LOC108348144
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 |
| chr10_+_764421 | 4.23 |
ENSRNOT00000084608
ENSRNOT00000087567 |
Myh11
|
myosin heavy chain 11 |
| chr14_-_8510138 | 4.23 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chrM_+_7758 | 4.19 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr4_-_22425381 | 4.17 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chrM_+_7919 | 4.17 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr17_+_69654170 | 4.11 |
ENSRNOT00000058351
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr4_-_171176581 | 4.07 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chrX_-_158978995 | 3.97 |
ENSRNOT00000001179
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr16_+_27399467 | 3.96 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chrX_+_68771100 | 3.88 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr1_-_141655417 | 3.86 |
ENSRNOT00000068084
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr18_-_58423196 | 3.66 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr13_+_98311827 | 3.61 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr15_+_11298478 | 3.57 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr17_-_77527894 | 3.55 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr2_-_9472384 | 3.55 |
ENSRNOT00000084295
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr7_-_120027026 | 3.52 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr2_-_35104963 | 3.43 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr17_-_48810991 | 3.43 |
ENSRNOT00000067636
|
Vps41
|
VPS41 HOPS complex subunit |
| chr12_+_41486076 | 3.28 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr3_+_38559914 | 3.25 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr15_-_60752106 | 3.24 |
ENSRNOT00000058148
|
Akap11
|
A-kinase anchoring protein 11 |
| chr18_+_58325319 | 3.20 |
ENSRNOT00000065802
ENSRNOT00000077726 |
Napg
|
NSF attachment protein gamma |
| chr17_-_9441526 | 3.01 |
ENSRNOT00000000146
|
Txndc15
|
thioredoxin domain containing 15 |
| chr18_-_26656879 | 2.99 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr5_-_78324278 | 2.90 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr1_-_141655894 | 2.88 |
ENSRNOT00000082591
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr3_-_92933725 | 2.85 |
ENSRNOT00000009552
|
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr2_-_140464607 | 2.79 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr4_-_112707270 | 2.71 |
ENSRNOT00000009308
|
Mrpl19
|
mitochondrial ribosomal protein L19 |
| chr3_+_8958090 | 2.69 |
ENSRNOT00000086789
ENSRNOT00000064557 |
Dolpp1
|
dolichyldiphosphatase 1 |
| chrX_-_23139694 | 2.67 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_-_190965115 | 2.61 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr19_-_9765484 | 2.53 |
ENSRNOT00000016626
|
Setd6
|
SET domain containing 6 |
| chr19_-_21970103 | 2.49 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr12_-_6078411 | 2.48 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr14_+_1355004 | 2.48 |
ENSRNOT00000043131
|
AABR07014114.1
|
|
| chr12_+_15890170 | 2.45 |
ENSRNOT00000001654
|
Gna12
|
G protein subunit alpha 12 |
| chrX_+_6273733 | 2.45 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr3_-_77227036 | 2.41 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr2_-_181531978 | 2.37 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr4_+_35279063 | 2.36 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chrX_+_124631881 | 2.32 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr5_-_28130803 | 2.31 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr14_+_104480662 | 2.31 |
ENSRNOT00000078710
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr3_+_16241573 | 2.29 |
ENSRNOT00000082831
|
Olr407
|
olfactory receptor 407 |
| chr13_-_91020112 | 2.27 |
ENSRNOT00000030449
|
Dusp23
|
dual specificity phosphatase 23 |
| chr1_+_67025240 | 2.27 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr1_-_15374850 | 2.25 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_+_140477868 | 2.18 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chrX_-_63291107 | 2.15 |
ENSRNOT00000092019
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr1_-_64147251 | 2.11 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr6_+_107603580 | 2.06 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr10_+_31813814 | 2.04 |
ENSRNOT00000009573
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chrX_-_63291371 | 2.03 |
ENSRNOT00000082421
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr5_-_99033107 | 1.96 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr3_-_45169118 | 1.94 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr13_+_85465792 | 1.94 |
ENSRNOT00000082991
ENSRNOT00000005277 |
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr8_-_43796224 | 1.92 |
ENSRNOT00000084315
|
Olr1330
|
olfactory receptor 1330 |
| chr4_-_129619142 | 1.91 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr2_-_199354793 | 1.91 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr4_+_25635765 | 1.85 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr12_+_10574168 | 1.84 |
ENSRNOT00000092401
|
Rnf6
|
ring finger protein 6 |
| chr3_-_37803112 | 1.79 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr1_+_249574954 | 1.78 |
ENSRNOT00000074100
|
Cstf2t
|
cleavage stimulation factor subunit 2, tau variant |
| chr8_+_19972793 | 1.76 |
ENSRNOT00000068066
|
Olr1159
|
olfactory receptor 1159 |
| chr15_+_28023018 | 1.75 |
ENSRNOT00000090272
|
Rnase4
|
ribonuclease A family member 4 |
| chr15_-_33576331 | 1.75 |
ENSRNOT00000021851
|
Slc22a17
|
solute carrier family 22, member 17 |
| chr20_-_5754773 | 1.73 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr16_+_18716019 | 1.73 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr7_-_12831675 | 1.73 |
ENSRNOT00000011936
|
Fgf22
|
fibroblast growth factor 22 |
| chr10_-_12312692 | 1.71 |
ENSRNOT00000073059
|
Olr1355
|
olfactory receptor 1355 |
| chr8_-_18037774 | 1.71 |
ENSRNOT00000074948
|
Olr1137
|
olfactory receptor 1137 |
| chr4_+_121778227 | 1.68 |
ENSRNOT00000084689
|
Vom1r93
|
vomeronasal 1 receptor 93 |
| chr1_+_252409268 | 1.67 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr1_-_140477796 | 1.67 |
ENSRNOT00000067172
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr18_+_61563053 | 1.62 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chr7_-_9104719 | 1.56 |
ENSRNOT00000088247
|
LOC103692758
|
olfactory receptor 6C74-like |
| chr2_+_243018975 | 1.56 |
ENSRNOT00000081989
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr8_+_99880073 | 1.53 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr3_-_77800137 | 1.52 |
ENSRNOT00000071319
|
AABR07052831.1
|
|
| chr5_+_113725717 | 1.51 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr19_-_24487396 | 1.46 |
ENSRNOT00000031614
|
Elmod2
|
ELMO domain containing 2 |
| chr15_+_26887767 | 1.44 |
ENSRNOT00000016674
|
Olr1627
|
olfactory receptor 1627 |
| chr3_+_73307900 | 1.40 |
ENSRNOT00000051690
|
Olr469
|
olfactory receptor 469 |
| chr12_-_19592000 | 1.36 |
ENSRNOT00000001855
|
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr2_+_144522072 | 1.35 |
ENSRNOT00000057978
|
Spg20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr20_+_32717564 | 1.35 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr5_-_130085838 | 1.32 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr7_+_42304534 | 1.31 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr12_+_4310334 | 1.31 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr14_-_10749120 | 1.28 |
ENSRNOT00000003046
|
Cops4
|
COP9 signalosome subunit 4 |
| chr13_-_35511020 | 1.27 |
ENSRNOT00000003413
|
Ralb
|
RAS like proto-oncogene B |
| chr3_-_77826506 | 1.26 |
ENSRNOT00000071385
|
Olr675
|
olfactory receptor 675 |
| chr19_+_27603307 | 1.23 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chrX_-_83864150 | 1.20 |
ENSRNOT00000073734
|
Obp1f
|
odorant binding protein I f |
| chr1_-_168611670 | 1.19 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr11_+_31893489 | 1.18 |
ENSRNOT00000042750
|
Itsn1
|
intersectin 1 |
| chr3_+_112390106 | 1.18 |
ENSRNOT00000081197
|
LOC691918
|
similar to Centrosomal protein of 27 kDa (Cep27 protein) |
| chrX_+_22342308 | 1.17 |
ENSRNOT00000079909
ENSRNOT00000087945 ENSRNOT00000077528 |
Kdm5c
|
lysine demethylase 5C |
| chr19_+_43338166 | 1.17 |
ENSRNOT00000091720
ENSRNOT00000084739 |
Fuk
|
fucokinase |
| chr14_-_11198194 | 1.17 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr3_-_78235760 | 1.15 |
ENSRNOT00000008435
|
Olr698
|
olfactory receptor 698 |
| chr10_+_34756094 | 1.14 |
ENSRNOT00000050554
|
Olr1399
|
olfactory receptor 1399 |
| chr4_-_88684415 | 1.13 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr3_-_73401074 | 1.12 |
ENSRNOT00000012848
|
Olr476
|
olfactory receptor 476 |
| chr4_-_167089055 | 1.11 |
ENSRNOT00000050409
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr6_+_3012804 | 1.07 |
ENSRNOT00000061980
|
Arhgef33
|
Rho guanine nucleotide exchange factor 33 |
| chr10_+_15484795 | 1.06 |
ENSRNOT00000079826
|
Tmem8a
|
transmembrane protein 8A |
| chr1_-_71486790 | 1.04 |
ENSRNOT00000041542
|
Nlrp5
|
NLR family, pyrin domain containing 5 |
| chr6_-_76552559 | 1.02 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr1_-_172076138 | 1.00 |
ENSRNOT00000051241
|
Olr239
|
olfactory receptor 239 |
| chr8_+_19548488 | 1.00 |
ENSRNOT00000008605
|
Olr1148
|
olfactory receptor 1148 |
| chr3_+_175708519 | 0.99 |
ENSRNOT00000085983
|
AC115322.1
|
|
| chr2_-_96509424 | 0.97 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr19_+_33928356 | 0.97 |
ENSRNOT00000058162
|
Ednra
|
endothelin receptor type A |
| chr3_-_73677403 | 0.95 |
ENSRNOT00000084063
|
Olr496
|
olfactory receptor 496 |
| chrX_+_45213228 | 0.93 |
ENSRNOT00000088538
|
LOC103690319
|
odorant-binding protein |
| chr16_-_75156321 | 0.88 |
ENSRNOT00000058063
|
Defb11
|
defensin beta 11 |
| chr6_-_1454480 | 0.86 |
ENSRNOT00000072810
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_+_177013604 | 0.84 |
ENSRNOT00000020581
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr8_+_132135187 | 0.84 |
ENSRNOT00000091791
|
Tgm4
|
transglutaminase 4 |
| chr8_-_126390801 | 0.83 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr16_-_75208230 | 0.82 |
ENSRNOT00000058059
|
Defb10
|
defensin beta 10 |
| chr17_-_63879166 | 0.78 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr7_+_54859326 | 0.77 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr8_+_107229832 | 0.74 |
ENSRNOT00000045821
|
Faim
|
Fas apoptotic inhibitory molecule |
| chrX_-_45284341 | 0.72 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr1_-_190370499 | 0.70 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr20_+_31234493 | 0.65 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr1_-_172665244 | 0.62 |
ENSRNOT00000089570
|
Olr264
|
olfactory receptor 264 |
| chr2_-_188559882 | 0.57 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr7_+_6215743 | 0.57 |
ENSRNOT00000050967
|
Olr1002
|
olfactory receptor 1002 |
| chr4_+_166076156 | 0.56 |
ENSRNOT00000007124
|
Prp2
|
proline rich protein 2 |
| chr3_+_15895191 | 0.55 |
ENSRNOT00000047879
|
Olr400
|
olfactory receptor 400 |
| chr1_+_168411463 | 0.54 |
ENSRNOT00000072397
|
Olr89
|
olfactory receptor 89 |
| chr17_+_36691875 | 0.54 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr8_+_133285064 | 0.54 |
ENSRNOT00000063989
|
B230307C23Rik
|
RIKEN cDNA B230307C23 gene |
| chr1_+_213258684 | 0.53 |
ENSRNOT00000091678
|
LOC103690271
|
olfactory receptor 13A1-like |
| chr7_+_73273985 | 0.53 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr7_-_17859653 | 0.51 |
ENSRNOT00000030164
|
Zscan4f
|
zinc finger and SCAN domain containing 4F |
| chr11_-_88374679 | 0.50 |
ENSRNOT00000002524
|
Top3b
|
topoisomerase (DNA) III beta |
| chr1_-_170694872 | 0.50 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr13_+_79378733 | 0.45 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr8_+_106816152 | 0.45 |
ENSRNOT00000047613
|
Faim
|
Fas apoptotic inhibitory molecule |
| chr3_-_125470413 | 0.45 |
ENSRNOT00000028893
|
Trmt6
|
tRNA methyltransferase 6 |
| chr10_-_27427686 | 0.44 |
ENSRNOT00000087475
|
AABR07029410.1
|
|
| chr1_-_229927937 | 0.42 |
ENSRNOT00000080145
|
Olr349
|
olfactory receptor 349 |
| chr1_-_168930776 | 0.40 |
ENSRNOT00000021468
|
LOC103694854
|
olfactory receptor 52Z1-like |
| chr2_-_149088787 | 0.40 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr1_+_261180007 | 0.39 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr3_+_44025300 | 0.34 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0016488 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 5.5 | 16.4 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 5.2 | 10.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 3.9 | 11.7 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 3.3 | 10.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 3.3 | 13.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 3.3 | 10.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 3.2 | 9.7 | GO:0006742 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 2.9 | 8.6 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 2.6 | 10.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.3 | 6.9 | GO:0060769 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 2.0 | 6.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.5 | 4.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.2 | 5.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 1.2 | 7.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.0 | 4.2 | GO:1905235 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.7 | 3.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.7 | 9.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.7 | 2.0 | GO:0097214 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) lysosomal membrane organization(GO:0097212) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.6 | 1.9 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.6 | 5.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.6 | 1.7 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 4.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.6 | 4.5 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.6 | 10.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.5 | 3.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 1.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 1.4 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.4 | 1.3 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.4 | 4.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 11.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.4 | 5.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.4 | 2.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 2.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 5.3 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.4 | 1.2 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.4 | 7.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 1.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.4 | 3.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 2.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.3 | 2.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.3 | 4.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 6.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 2.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 2.4 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.3 | 2.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 8.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 5.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 13.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 3.8 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 7.6 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.3 | 1.3 | GO:0070666 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 2.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 7.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 3.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 4.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 2.9 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 1.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 1.0 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 1.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 0.8 | GO:0042628 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.2 | 0.5 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 1.9 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 2.3 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 3.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.9 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.1 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 3.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 2.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 1.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 3.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 1.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 3.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 7.0 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 4.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 2.4 | GO:0043487 | regulation of RNA stability(GO:0043487) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 1.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 4.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 24.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 2.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 3.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 2.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.6 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 2.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.9 | 9.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.7 | 5.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 7.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 4.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.7 | 10.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.7 | 8.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.7 | 2.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.6 | 3.5 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.6 | 10.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 8.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.5 | 4.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 4.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 5.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.3 | 10.5 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 2.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 5.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 16.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 7.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 13.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.9 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 5.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 7.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 5.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 7.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 4.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 8.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 63.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 6.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.7 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 4.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 13.9 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 4.4 | 13.3 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 3.4 | 10.3 | GO:0070401 | androsterone dehydrogenase activity(GO:0047023) NADP+ binding(GO:0070401) |
| 3.4 | 10.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 3.3 | 16.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 2.4 | 9.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 2.3 | 7.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 2.0 | 6.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.9 | 11.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.1 | 3.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.0 | 9.3 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 1.0 | 4.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.9 | 6.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.8 | 4.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.8 | 2.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.7 | 2.9 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.6 | 4.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 4.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 5.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 2.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.5 | 2.4 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.4 | 3.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 5.3 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.4 | 2.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 7.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 2.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 1.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 2.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 7.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 10.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 13.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 10.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.2 | 1.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 3.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.5 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 5.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 1.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 4.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 13.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 6.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 1.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 8.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 8.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 4.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 2.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 8.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 6.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 10.0 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 3.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 13.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 5.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 8.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 17.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 5.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.8 | GO:0004519 | endonuclease activity(GO:0004519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 10.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 6.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 5.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 5.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 10.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 4.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 6.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 2.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 6.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 18.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 4.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 19.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 6.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 4.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 9.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |