Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
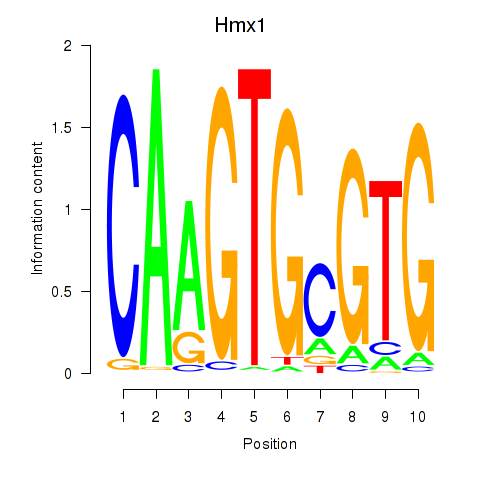
Results for Hmx1
Z-value: 0.71
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSRNOG00000009154 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx1 | rn6_v1_chr14_-_80544342_80544342 | -0.39 | 7.1e-13 | Click! |
Activity profile of Hmx1 motif
Sorted Z-values of Hmx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_169658875 | 24.23 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr10_-_31359699 | 21.72 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_+_20607507 | 19.15 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr15_-_35113678 | 16.64 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr15_-_61564695 | 16.15 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr7_-_119797098 | 15.60 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr5_+_148193710 | 14.96 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr6_-_92760018 | 13.16 |
ENSRNOT00000009560
|
Trim9
|
tripartite motif-containing 9 |
| chr8_+_33816386 | 13.06 |
ENSRNOT00000011925
|
Ets1
|
ETS proto-oncogene 1, transcription factor |
| chr7_-_121058029 | 13.00 |
ENSRNOT00000068033
|
Cbx6
|
chromobox 6 |
| chr5_+_152681101 | 13.00 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr5_-_77173502 | 12.45 |
ENSRNOT00000023588
|
Slc46a2
|
solute carrier family 46, member 2 |
| chr5_+_1417478 | 11.44 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr10_-_31419235 | 11.08 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_-_93307420 | 10.76 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr7_+_133856101 | 10.40 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr17_-_9791781 | 9.88 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr7_-_140483693 | 9.85 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr1_-_104202591 | 9.46 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr7_+_120140460 | 9.34 |
ENSRNOT00000040513
ENSRNOT00000073905 |
Pdxp
|
pyridoxal phosphatase |
| chr17_+_33408722 | 9.19 |
ENSRNOT00000023691
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr17_-_9792007 | 9.04 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr13_+_31081804 | 8.91 |
ENSRNOT00000041413
|
Cdh7
|
cadherin 7 |
| chr10_+_47281786 | 8.56 |
ENSRNOT00000089123
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr2_-_187909394 | 8.32 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chr15_-_60512704 | 8.23 |
ENSRNOT00000049056
|
Tnfsf11
|
tumor necrosis factor superfamily member 11 |
| chr9_+_67763897 | 8.14 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chr14_+_75852060 | 8.07 |
ENSRNOT00000075975
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr8_-_53899669 | 8.03 |
ENSRNOT00000082257
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr10_-_108691367 | 7.62 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr10_-_13004441 | 7.49 |
ENSRNOT00000004848
|
Cldn9
|
claudin 9 |
| chr4_-_157252104 | 7.40 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr10_+_4957326 | 7.35 |
ENSRNOT00000003458
|
Socs1
|
suppressor of cytokine signaling 1 |
| chr10_-_4910305 | 7.29 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr3_+_122803772 | 7.12 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chrX_+_138046494 | 7.07 |
ENSRNOT00000010596
|
Stk26
|
serine/threonine kinase 26 |
| chr13_+_47602692 | 6.88 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr20_+_5509059 | 6.79 |
ENSRNOT00000065349
|
Kifc1
|
kinesin family member C1 |
| chr4_+_56711049 | 6.70 |
ENSRNOT00000027237
|
Flnc
|
filamin C |
| chr9_-_46401911 | 6.58 |
ENSRNOT00000046557
|
Creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr15_+_3938075 | 6.56 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr4_+_56711275 | 6.52 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr13_+_52553775 | 6.43 |
ENSRNOT00000011991
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr1_-_89360733 | 6.34 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr8_+_117347029 | 6.11 |
ENSRNOT00000047717
|
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr10_+_3411380 | 5.98 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr2_+_69415057 | 5.96 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr12_+_41385241 | 5.91 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr13_+_70174936 | 5.88 |
ENSRNOT00000064068
ENSRNOT00000079861 ENSRNOT00000092562 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chr18_+_87415531 | 5.83 |
ENSRNOT00000072475
|
AABR07032888.1
|
|
| chr2_-_25095106 | 5.80 |
ENSRNOT00000051653
ENSRNOT00000085857 |
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr10_+_90731865 | 5.69 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr3_-_141411170 | 5.41 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr3_-_175327689 | 5.30 |
ENSRNOT00000082981
|
Taf4
|
TATA-box binding protein associated factor 4 |
| chr10_+_90731148 | 5.26 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr7_+_119647375 | 5.20 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr8_-_33017854 | 5.07 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr1_-_2017488 | 4.98 |
ENSRNOT00000021905
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr2_-_188553289 | 4.98 |
ENSRNOT00000088822
|
Trim46
|
tripartite motif-containing 46 |
| chr10_-_85574889 | 4.94 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr2_+_211381036 | 4.64 |
ENSRNOT00000055862
|
Wdr47
|
WD repeat domain 47 |
| chr2_+_211078334 | 4.55 |
ENSRNOT00000049127
|
Sort1
|
sortilin 1 |
| chr7_-_70969905 | 4.50 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr8_-_49045154 | 4.39 |
ENSRNOT00000088034
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr20_-_45815940 | 4.38 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chr8_-_22336794 | 4.29 |
ENSRNOT00000066340
|
Ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr20_+_3367208 | 4.29 |
ENSRNOT00000091149
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr3_+_110918243 | 4.22 |
ENSRNOT00000056432
|
Rad51
|
RAD51 recombinase |
| chr3_-_122947075 | 4.22 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_+_50828585 | 4.20 |
ENSRNOT00000086441
ENSRNOT00000023290 ENSRNOT00000035402 |
LOC108348175
|
protein quaking-like |
| chrX_+_14498119 | 4.18 |
ENSRNOT00000051135
|
Xk
|
X-linked Kx blood group |
| chr15_+_36809361 | 4.18 |
ENSRNOT00000076667
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr19_+_38039564 | 4.05 |
ENSRNOT00000087491
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr7_+_145117951 | 4.00 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr10_+_55275411 | 3.97 |
ENSRNOT00000065895
|
Myh10
|
myosin heavy chain 10 |
| chr3_+_148579920 | 3.85 |
ENSRNOT00000012432
|
Hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_-_47724499 | 3.82 |
ENSRNOT00000085011
|
Rnf112
|
ring finger protein 112 |
| chr20_+_12429315 | 3.79 |
ENSRNOT00000001675
|
Pcbp3
|
poly(rC) binding protein 3 |
| chr1_-_67390141 | 3.78 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chr10_+_48240330 | 3.73 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chrX_+_37469937 | 3.72 |
ENSRNOT00000008938
|
Rps6ka3
|
ribosomal protein S6 kinase A3 |
| chrX_+_156463953 | 3.61 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr1_+_50828134 | 3.60 |
ENSRNOT00000089149
|
LOC108348175
|
protein quaking-like |
| chrX_+_14498283 | 3.55 |
ENSRNOT00000092143
|
Xk
|
X-linked Kx blood group |
| chr5_-_150439959 | 3.55 |
ENSRNOT00000014506
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr18_+_31094965 | 3.45 |
ENSRNOT00000026526
|
Rell2
|
RELT-like 2 |
| chr8_+_114866768 | 3.23 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr7_+_120380544 | 3.19 |
ENSRNOT00000015009
|
Polr2f
|
RNA polymerase II subunit F |
| chr11_-_86303453 | 3.17 |
ENSRNOT00000071453
|
LOC498122
|
similar to CG15908-PA |
| chr6_+_137184820 | 3.15 |
ENSRNOT00000073796
|
Adssl1
|
adenylosuccinate synthase like 1 |
| chr12_+_13090172 | 3.09 |
ENSRNOT00000092558
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr1_-_165680176 | 3.09 |
ENSRNOT00000025245
ENSRNOT00000082697 |
Plekhb1
|
pleckstrin homology domain containing B1 |
| chr19_+_37252843 | 3.09 |
ENSRNOT00000021145
|
E2f4
|
E2F transcription factor 4 |
| chr8_+_103774358 | 3.05 |
ENSRNOT00000014481
|
Xrn1
|
5'-3' exoribonuclease 1 |
| chr12_+_13090367 | 3.01 |
ENSRNOT00000001417
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr8_+_114867062 | 2.96 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr17_+_26785029 | 2.93 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr5_-_113532878 | 2.88 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr4_+_61912210 | 2.85 |
ENSRNOT00000013569
|
Bpgm
|
bisphosphoglycerate mutase |
| chr4_+_58053041 | 2.85 |
ENSRNOT00000072698
|
Mest
|
mesoderm specific transcript |
| chr3_+_11921715 | 2.82 |
ENSRNOT00000021689
|
Fam129b
|
family with sequence similarity 129, member B |
| chr10_+_104523996 | 2.68 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr8_+_19888667 | 2.65 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr6_-_26792808 | 2.63 |
ENSRNOT00000010403
|
Abhd1
|
abhydrolase domain containing 1 |
| chr18_+_29999290 | 2.56 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_190370499 | 2.45 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr1_-_231530690 | 2.31 |
ENSRNOT00000087140
ENSRNOT00000018452 ENSRNOT00000018272 |
Tle4
|
transducin-like enhancer of split 4 |
| chr1_-_261446570 | 2.27 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr12_+_17735740 | 2.21 |
ENSRNOT00000001775
|
Pdgfa
|
platelet derived growth factor subunit A |
| chr5_-_109621170 | 2.19 |
ENSRNOT00000093007
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr20_+_4357733 | 2.19 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr10_-_90265017 | 2.02 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr7_+_11737293 | 1.96 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr13_+_57131395 | 1.96 |
ENSRNOT00000017884
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr7_-_140454268 | 1.94 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr1_-_262013619 | 1.78 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr5_+_135074297 | 1.74 |
ENSRNOT00000000157
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr3_+_2490518 | 1.61 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_+_221409271 | 1.58 |
ENSRNOT00000028462
|
Syvn1
|
synoviolin 1 |
| chr4_+_130172727 | 1.49 |
ENSRNOT00000051121
|
Mitf
|
melanogenesis associated transcription factor |
| chr15_-_35394792 | 1.48 |
ENSRNOT00000028058
|
Gzmc
|
granzyme C |
| chr1_-_261229046 | 1.48 |
ENSRNOT00000075531
|
Mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr4_+_157374318 | 1.48 |
ENSRNOT00000071027
|
AC115420.2
|
|
| chr4_+_140703619 | 1.36 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr5_+_48374096 | 1.34 |
ENSRNOT00000010172
|
Gabrr1
|
gamma-aminobutyric acid type A receptor rho 1 subunit |
| chr18_+_30004565 | 1.33 |
ENSRNOT00000027393
|
Pcdha4
|
protocadherin alpha 4 |
| chr5_+_149077412 | 1.16 |
ENSRNOT00000014666
|
Matn1
|
matrilin 1, cartilage matrix protein |
| chr8_+_67295727 | 1.06 |
ENSRNOT00000020443
|
Anp32a
|
acidic nuclear phosphoprotein 32 family member A |
| chr2_-_197935567 | 1.03 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr14_-_12387102 | 1.02 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr16_-_3912148 | 0.93 |
ENSRNOT00000014941
|
Anxa11
|
annexin A11 |
| chr15_+_93634820 | 0.82 |
ENSRNOT00000093318
|
Cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr1_-_59732409 | 0.78 |
ENSRNOT00000014824
|
Has1
|
hyaluronan synthase 1 |
| chr8_-_39437270 | 0.77 |
ENSRNOT00000076158
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr6_+_10533151 | 0.77 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr10_+_43224269 | 0.76 |
ENSRNOT00000003473
|
Sap30l
|
SAP30-like |
| chr10_-_109802739 | 0.76 |
ENSRNOT00000054951
|
Sirt7
|
sirtuin 7 |
| chr4_+_171730635 | 0.75 |
ENSRNOT00000009923
|
Strap
|
serine/threonine kinase receptor associated protein |
| chr10_+_103874383 | 0.75 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr9_+_94324793 | 0.70 |
ENSRNOT00000092493
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr15_+_33333420 | 0.66 |
ENSRNOT00000049927
|
RGD1308430
|
similar to 1700123O20Rik protein |
| chr3_-_171134655 | 0.60 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr20_-_12128747 | 0.52 |
ENSRNOT00000001644
|
Pofut2
|
protein O-fucosyltransferase 2 |
| chr7_-_3123297 | 0.46 |
ENSRNOT00000061878
|
Rab5b
|
RAB5B, member RAS oncogene family |
| chr18_+_29993361 | 0.46 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_-_143931940 | 0.38 |
ENSRNOT00000086214
|
Exosc8
|
exosome component 8 |
| chr2_+_57206613 | 0.34 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr12_-_17519954 | 0.33 |
ENSRNOT00000089418
|
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr19_+_24044103 | 0.32 |
ENSRNOT00000004621
|
Rnf150
|
ring finger protein 150 |
| chr12_-_11808977 | 0.30 |
ENSRNOT00000071795
|
AABR07035375.1
|
|
| chr17_+_18151982 | 0.24 |
ENSRNOT00000066447
|
Kif13a
|
kinesin family member 13A |
| chr15_+_86153628 | 0.20 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr8_-_118182559 | 0.18 |
ENSRNOT00000084838
|
Dhx30
|
DExH-box helicase 30 |
| chr3_-_176926722 | 0.18 |
ENSRNOT00000020168
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr13_+_52147555 | 0.16 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr15_-_36472327 | 0.14 |
ENSRNOT00000032601
ENSRNOT00000059660 |
LOC102553861
|
granzyme-like protein 1-like |
| chr4_-_159697207 | 0.09 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr1_-_262014066 | 0.04 |
ENSRNOT00000087083
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr15_-_35417273 | 0.03 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.9 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 5.5 | 16.6 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 5.4 | 16.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 4.4 | 13.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 4.1 | 32.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 3.3 | 13.1 | GO:0030578 | PML body organization(GO:0030578) |
| 3.2 | 13.0 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 3.1 | 9.3 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 3.1 | 9.2 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 2.7 | 8.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 2.4 | 24.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 2.3 | 4.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 2.1 | 8.2 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of osteoclast development(GO:2001206) |
| 1.8 | 7.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.8 | 5.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.6 | 9.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.5 | 6.0 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.4 | 19.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.4 | 21.7 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 1.3 | 4.0 | GO:0021678 | third ventricle development(GO:0021678) |
| 1.3 | 7.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 1.2 | 6.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.1 | 4.2 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 1.1 | 3.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.9 | 4.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 2.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 2.2 | GO:0060683 | regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling(GO:0060683) embryonic lung development(GO:1990401) |
| 0.7 | 3.6 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.7 | 7.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.7 | 2.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.7 | 5.9 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.6 | 1.9 | GO:0051884 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) anagen(GO:0042640) regulation of anagen(GO:0051884) |
| 0.6 | 6.9 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.6 | 7.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.6 | 7.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.5 | 6.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.4 | 8.1 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.4 | 8.3 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.4 | 0.8 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.4 | 2.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 12.4 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.4 | 4.0 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.4 | 6.3 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.4 | 3.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.4 | 1.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.3 | 4.4 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 5.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 6.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.3 | 2.7 | GO:0061450 | nail development(GO:0035878) trophoblast cell migration(GO:0061450) |
| 0.3 | 2.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.3 | 4.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 8.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 7.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.3 | 4.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 20.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 5.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 3.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 3.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 5.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 7.1 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.2 | 13.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.2 | 0.8 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 1.2 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 5.9 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.2 | 6.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 1.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 5.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 3.8 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.1 | 19.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 11.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 7.3 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 2.8 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 6.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.3 | GO:0030473 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 1.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 3.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 4.2 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 2.9 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 2.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.9 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 1.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 7.5 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.0 | 5.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 3.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.0 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 2.1 | 6.3 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 1.5 | 24.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.0 | 7.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.0 | 5.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.0 | 4.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.9 | 7.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.7 | 8.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.7 | 9.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.7 | 9.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 8.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.5 | 1.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 6.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 5.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 5.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.4 | 1.8 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.3 | 16.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 13.2 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 1.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 4.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 2.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 3.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 6.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 2.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 23.1 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 6.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 22.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 8.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 4.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 11.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 8.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 7.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 10.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 5.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 20.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 4.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 30.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 17.7 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 4.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 10.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 26.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.8 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 4.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 5.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 4.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 6.2 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.8 | 9.2 | GO:0070401 | NADP+ binding(GO:0070401) |
| 1.3 | 8.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.3 | 24.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 1.3 | 18.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 1.1 | 4.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.0 | 7.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.0 | 4.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.8 | 7.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 0.8 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.8 | 3.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.7 | 2.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.7 | 4.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.7 | 6.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.7 | 5.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.6 | 3.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 2.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 16.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 6.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 4.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 3.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.4 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 3.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.4 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.4 | 8.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 13.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 7.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.3 | 7.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 8.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 5.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 4.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 2.0 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 6.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 5.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 4.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 6.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 5.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.8 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 1.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 23.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 5.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 4.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 5.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 9.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 3.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 12.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 13.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 7.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 9.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 4.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 6.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.1 | 6.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 7.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 9.3 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 4.3 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.6 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 6.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 4.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 11.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 2.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 44.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.6 | 6.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 19.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.5 | 3.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 16.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 27.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.4 | 16.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 7.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 9.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 3.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 8.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 10.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 6.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 2.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.8 | 13.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.8 | 9.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 25.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 14.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.5 | 4.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 14.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 24.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 5.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 5.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 8.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 8.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 6.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 7.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 6.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 3.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 3.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 5.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 8.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 5.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 3.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 4.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 8.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 4.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 13.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |