Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hmga2
Z-value: 0.95
Transcription factors associated with Hmga2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmga2
|
ENSRNOG00000042460 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga2 | rn6_v1_chr7_-_65275408_65275408 | -0.21 | 2.0e-04 | Click! |
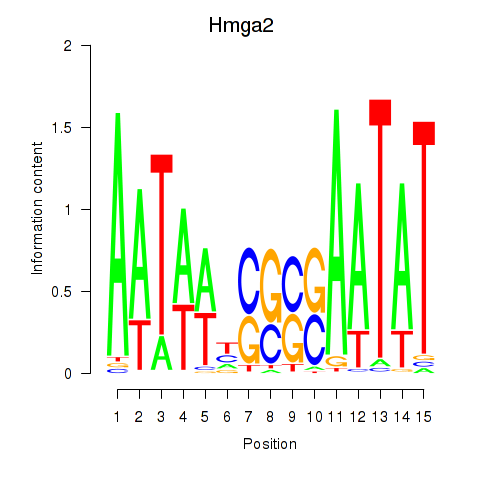
Activity profile of Hmga2 motif
Sorted Z-values of Hmga2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_55219773 | 46.79 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr2_+_74360622 | 24.43 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr1_-_148119857 | 21.59 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr6_+_113898420 | 20.48 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr19_+_209926 | 19.98 |
ENSRNOT00000071393
|
LOC108348293
|
uncharacterized LOC108348293 |
| chr8_+_41657566 | 19.82 |
ENSRNOT00000042860
|
Rup2
|
urinary protein 2 |
| chr4_+_70776046 | 19.30 |
ENSRNOT00000040403
|
Prss1
|
protease, serine 1 |
| chr1_-_258766881 | 18.79 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr8_-_36760742 | 18.23 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr4_-_70628470 | 18.10 |
ENSRNOT00000029319
|
Try5
|
trypsin 5 |
| chr3_+_159823878 | 17.90 |
ENSRNOT00000011748
|
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1 |
| chrX_+_28072826 | 17.87 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr4_+_70614524 | 17.49 |
ENSRNOT00000041100
|
Prss3
|
protease, serine 3 |
| chr1_-_258877045 | 16.83 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr15_+_44799334 | 16.74 |
ENSRNOT00000018599
|
Nefl
|
neurofilament light |
| chr3_-_37854561 | 16.74 |
ENSRNOT00000076095
|
Neb
|
nebulin |
| chr6_-_23316962 | 15.38 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_+_32973695 | 15.21 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr11_+_36851038 | 15.16 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr10_+_96639924 | 15.06 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr9_-_73958480 | 15.01 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr3_-_102151489 | 14.24 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr14_+_22375955 | 14.16 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr11_-_4332255 | 14.01 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr10_-_103848035 | 13.99 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr2_+_154921999 | 13.94 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chr18_+_83471342 | 13.66 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr16_+_68586235 | 13.55 |
ENSRNOT00000039592
|
LOC103693984
|
uncharacterized LOC103693984 |
| chr1_-_102780381 | 12.97 |
ENSRNOT00000080132
|
Saa4
|
serum amyloid A4 |
| chrX_+_122808605 | 12.96 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr3_-_48831417 | 12.94 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr16_+_35573058 | 12.76 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr18_-_6781841 | 12.73 |
ENSRNOT00000077606
ENSRNOT00000048109 |
Aqp4
|
aquaporin 4 |
| chr5_-_173233188 | 12.45 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr1_-_31055453 | 12.32 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr1_+_147713892 | 12.26 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr4_-_16654811 | 12.26 |
ENSRNOT00000008637
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr1_+_106998623 | 12.25 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr4_+_138269142 | 12.24 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr5_-_146446227 | 11.47 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr2_+_93758919 | 11.42 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr4_-_27638676 | 11.41 |
ENSRNOT00000011377
|
RGD1306626
|
similar to RIKEN cDNA 4930500J03 |
| chr9_+_12346117 | 10.80 |
ENSRNOT00000074599
|
AABR07066677.1
|
|
| chrX_+_121612952 | 10.77 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr9_+_24095751 | 10.73 |
ENSRNOT00000018177
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr12_+_47179664 | 10.46 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr3_-_25212049 | 10.42 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_+_248428099 | 10.41 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr10_-_103340922 | 10.35 |
ENSRNOT00000004191
|
Btbd17
|
BTB domain containing 17 |
| chr1_-_162713610 | 10.22 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr12_+_10636275 | 10.10 |
ENSRNOT00000001285
|
Cyp3a18
|
cytochrome P450, family 3, subfamily a, polypeptide 18 |
| chr1_+_107262659 | 10.06 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr3_-_94182714 | 9.91 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chr1_-_67302751 | 9.78 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr4_+_70755795 | 9.75 |
ENSRNOT00000043527
|
LOC683849
|
similar to Anionic trypsin II precursor (Pretrypsinogen II) |
| chr4_-_70747226 | 9.60 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr19_-_29373989 | 9.56 |
ENSRNOT00000078023
|
AABR07043564.1
|
|
| chr8_+_49418965 | 9.55 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr5_+_122508388 | 9.52 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr3_-_158328881 | 9.47 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr18_+_52215682 | 9.36 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr1_-_76517134 | 9.34 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr1_+_37507276 | 9.04 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr4_+_96831880 | 8.93 |
ENSRNOT00000068400
|
RSA-14-44
|
RSA-14-44 protein |
| chr5_-_166116516 | 8.92 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr7_-_14435967 | 8.82 |
ENSRNOT00000074801
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr9_+_12633990 | 8.66 |
ENSRNOT00000066517
ENSRNOT00000077532 |
Dazl
|
deleted in azoospermia-like |
| chr2_-_89310946 | 8.64 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr7_-_54855557 | 8.43 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr10_-_91291774 | 8.29 |
ENSRNOT00000004356
|
LOC100361655
|
rCG33642-like |
| chr4_+_52199416 | 8.17 |
ENSRNOT00000009537
|
Spam1
|
sperm adhesion molecule 1 |
| chr19_+_29320701 | 8.17 |
ENSRNOT00000049600
|
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr19_+_487723 | 8.08 |
ENSRNOT00000061734
|
Ces2j
|
carboxylesterase 2J |
| chr1_-_67206713 | 8.03 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chr8_+_81949877 | 7.74 |
ENSRNOT00000030221
|
LOC100360828
|
cAMP-regulated phosphoprotein 19-like |
| chr2_+_147006830 | 7.71 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr16_+_39144972 | 7.70 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr2_+_66940057 | 7.51 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr7_+_133856101 | 7.51 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr9_-_17065600 | 7.51 |
ENSRNOT00000025638
|
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr4_+_84423653 | 7.45 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr14_+_22517774 | 7.38 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr5_-_166133491 | 7.33 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr6_-_146099053 | 7.28 |
ENSRNOT00000007233
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr1_+_42169501 | 7.25 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr1_+_201913148 | 7.20 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr4_+_65112944 | 7.17 |
ENSRNOT00000083672
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chrX_+_43881246 | 7.11 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr3_+_143172686 | 7.08 |
ENSRNOT00000006869
|
Cst9l
|
cystatin 9-like |
| chr1_-_52495582 | 6.87 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chr1_+_256382791 | 6.79 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr17_-_57984036 | 6.73 |
ENSRNOT00000022389
|
Idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr1_-_67175736 | 6.66 |
ENSRNOT00000046788
|
Vom1r44
|
vomeronasal 1 receptor 44 |
| chr1_-_91526570 | 6.64 |
ENSRNOT00000056583
|
Wdr88
|
WD repeat domain 88 |
| chr2_-_231409496 | 6.61 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr16_+_39353283 | 6.60 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr16_+_39145230 | 6.56 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chrX_+_152885246 | 6.30 |
ENSRNOT00000087074
|
AABR07042326.3
|
|
| chr15_-_11812485 | 6.09 |
ENSRNOT00000030991
ENSRNOT00000007795 |
Nek10
|
NIMA-related kinase 10 |
| chr1_+_66898946 | 6.07 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr15_+_5783951 | 6.02 |
ENSRNOT00000061040
|
LOC305806
|
similar to glutaredoxin 1 (thioltransferase); glutaredoxin |
| chr14_+_100311173 | 5.76 |
ENSRNOT00000031275
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr4_+_45567573 | 5.66 |
ENSRNOT00000089824
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr3_-_162035614 | 5.66 |
ENSRNOT00000047890
|
Zfp663
|
zinc finger protein 663 |
| chr1_-_188713270 | 5.61 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr2_+_150146234 | 5.34 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr10_-_41546346 | 5.32 |
ENSRNOT00000058627
|
LOC102552882
|
uncharacterized LOC102552882 |
| chr18_-_40452456 | 5.26 |
ENSRNOT00000004747
|
Mospd4
|
motile sperm domain containing 4 |
| chr2_-_231409988 | 5.17 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr10_-_41491554 | 4.99 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr15_+_31209775 | 4.97 |
ENSRNOT00000079661
|
AABR07017804.1
|
|
| chr20_-_54269525 | 4.93 |
ENSRNOT00000037559
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr6_+_29977797 | 4.89 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr12_-_8418966 | 4.82 |
ENSRNOT00000085869
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr9_+_84569597 | 4.76 |
ENSRNOT00000020161
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_7322424 | 4.75 |
ENSRNOT00000066725
ENSRNOT00000081112 |
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chr16_+_12510827 | 4.74 |
ENSRNOT00000077763
|
AABR07024716.1
|
|
| chr13_+_71322759 | 4.68 |
ENSRNOT00000071815
|
Teddm1
|
transmembrane epididymal protein 1 |
| chrX_-_43878881 | 4.60 |
ENSRNOT00000066493
|
Cldn34d
|
claudin 34D |
| chr20_+_41083317 | 4.51 |
ENSRNOT00000000660
|
Tspyl1
|
TSPY-like 1 |
| chr1_+_229030233 | 4.49 |
ENSRNOT00000084503
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr10_+_49368314 | 4.34 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr3_+_101010899 | 4.31 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr5_-_59553416 | 4.30 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr1_-_103426467 | 4.29 |
ENSRNOT00000045792
|
Mrgprb4
|
MAS-related GPR, member B4 |
| chr7_+_35309265 | 4.26 |
ENSRNOT00000080291
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr20_+_2515520 | 4.13 |
ENSRNOT00000001024
|
Rpp21
|
ribonuclease P/MRP 21 subunit |
| chr15_-_33263659 | 4.08 |
ENSRNOT00000018005
|
Psmb5
|
proteasome subunit beta 5 |
| chr18_+_30474947 | 4.03 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr1_-_54036068 | 4.02 |
ENSRNOT00000075019
|
RGD1560718
|
similar to putative protein kinase |
| chr14_-_21761317 | 3.98 |
ENSRNOT00000077006
|
2310003L06Rik
|
RIKEN cDNA 2310003L06 gene |
| chr1_-_83977236 | 3.96 |
ENSRNOT00000028434
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_51619875 | 3.74 |
ENSRNOT00000023319
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr18_+_3597240 | 3.73 |
ENSRNOT00000017045
|
RGD1311805
|
similar to RIKEN cDNA 2400010D15 |
| chr11_-_1983513 | 3.73 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chrX_+_90588124 | 3.70 |
ENSRNOT00000045685
|
Tgif2lx2
|
TGFB-induced factor homeobox 2-like, X-linked 2 |
| chr17_-_35130045 | 3.67 |
ENSRNOT00000089878
|
LOC100912478
|
vacuolar ATPase assembly integral membrane protein VMA21-like |
| chrX_+_97074710 | 3.60 |
ENSRNOT00000044379
|
RGD1561230
|
similar to RIKEN cDNA 4921511C20 gene |
| chr16_-_54628458 | 3.53 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr1_-_169334093 | 3.50 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr3_+_47439076 | 3.46 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr2_+_181331464 | 3.45 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chrX_+_74200972 | 3.39 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr4_-_176679815 | 3.37 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr1_-_275882444 | 3.34 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr17_-_57526461 | 3.31 |
ENSRNOT00000072723
|
LOC103690053
|
ankyrin repeat domain-containing protein 26-like |
| chr1_-_22596475 | 3.21 |
ENSRNOT00000021510
|
Taar1
|
trace-amine-associated receptor 1 |
| chr17_+_57983937 | 3.20 |
ENSRNOT00000022688
|
Wdr37
|
WD repeat domain 37 |
| chr1_-_66212418 | 3.11 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr15_+_30525754 | 3.08 |
ENSRNOT00000086842
|
AABR07017745.2
|
|
| chrX_+_74205842 | 3.06 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr3_-_73879647 | 3.00 |
ENSRNOT00000090000
|
Olr510
|
olfactory receptor 510 |
| chr1_+_168204985 | 2.99 |
ENSRNOT00000049036
|
Olr75
|
olfactory receptor 75 |
| chr17_+_5225835 | 2.96 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr10_+_13061170 | 2.95 |
ENSRNOT00000004936
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr11_-_74793673 | 2.94 |
ENSRNOT00000002338
ENSRNOT00000002343 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr7_-_11513415 | 2.92 |
ENSRNOT00000082486
|
Thop1
|
thimet oligopeptidase 1 |
| chr11_-_17684903 | 2.92 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr1_+_64928503 | 2.90 |
ENSRNOT00000086274
|
Vom2r80
|
vomeronasal 2 receptor, 80 |
| chr11_+_88047832 | 2.86 |
ENSRNOT00000046159
|
Ube2l3
|
ubiquitin-conjugating enzyme E2L 3 |
| chr3_+_75712483 | 2.86 |
ENSRNOT00000049264
|
Olr575
|
olfactory receptor 575 |
| chr18_+_31574821 | 2.84 |
ENSRNOT00000018499
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_+_35677984 | 2.84 |
ENSRNOT00000024609
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr1_-_60828260 | 2.83 |
ENSRNOT00000059671
|
Vom1r-ps112
|
vomeronasal 1 receptor pseudogene 112 |
| chr17_+_38929045 | 2.82 |
ENSRNOT00000081739
|
Prl7d1
|
prolactin family 7, subfamily d, member 1 |
| chr5_-_107447636 | 2.79 |
ENSRNOT00000040254
|
Ifna16l1
|
interferon, alpha 16-like 1 |
| chr2_-_35503638 | 2.76 |
ENSRNOT00000007560
|
LOC100910467
|
olfactory receptor 144-like |
| chr12_-_52658275 | 2.74 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr8_+_4019722 | 2.73 |
ENSRNOT00000073415
|
Vom2r23
|
vomeronasal 2 receptor, 23 |
| chr3_+_9822580 | 2.73 |
ENSRNOT00000010786
ENSRNOT00000093718 |
Usp20
|
ubiquitin specific peptidase 20 |
| chr8_-_13290773 | 2.70 |
ENSRNOT00000012217
|
RGD1561795
|
similar to RIKEN cDNA 1700012B09 |
| chr7_+_108613739 | 2.68 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chrX_+_144994139 | 2.67 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr3_+_43271596 | 2.64 |
ENSRNOT00000088589
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr15_+_86153628 | 2.64 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr15_-_29291454 | 2.61 |
ENSRNOT00000092212
|
AABR07017617.2
|
|
| chr1_-_172552099 | 2.55 |
ENSRNOT00000090373
|
Olr257
|
olfactory receptor 257 |
| chr18_+_22964210 | 2.54 |
ENSRNOT00000066816
|
Pik3c3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr2_-_170300408 | 2.52 |
ENSRNOT00000046681
|
Si
|
sucrase-isomaltase |
| chr9_-_86103158 | 2.49 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr4_-_165810121 | 2.44 |
ENSRNOT00000048210
|
Tas2r104
|
taste receptor, type 2, member 104 |
| chr20_+_1936438 | 2.43 |
ENSRNOT00000073981
|
Olr1749
|
olfactory receptor 1749 |
| chr3_-_72081079 | 2.41 |
ENSRNOT00000007914
|
Tmx2
|
thioredoxin-related transmembrane protein 2 |
| chr9_+_27546528 | 2.40 |
ENSRNOT00000051403
|
Khdc1
|
KH homology domain containing 1 |
| chr15_-_29345785 | 2.40 |
ENSRNOT00000075484
|
AABR07017623.1
|
|
| chr10_-_29367865 | 2.39 |
ENSRNOT00000005304
|
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr1_-_89084859 | 2.38 |
ENSRNOT00000032026
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr1_+_168341595 | 2.38 |
ENSRNOT00000048411
|
Olr84
|
olfactory receptor 84 |
| chr2_-_127754648 | 2.36 |
ENSRNOT00000087535
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr1_-_227486976 | 2.28 |
ENSRNOT00000035812
|
Ms4a14
|
membrane spanning 4-domains A14 |
| chr5_-_119505586 | 2.23 |
ENSRNOT00000089521
|
Cyp2j16
|
cytochrome P450, family 2, subfamily j, polypeptide 16 |
| chr1_+_79181855 | 2.22 |
ENSRNOT00000037132
|
Psgb1
|
pregnancy-specific beta 1-glycoprotein |
| chr5_+_157327657 | 2.21 |
ENSRNOT00000022847
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr18_+_40732480 | 2.21 |
ENSRNOT00000064116
|
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr17_+_39032661 | 2.19 |
ENSRNOT00000022381
|
Prl7a3
|
prolactin family 7, subfamily a, member 3 |
| chr3_+_16183322 | 2.19 |
ENSRNOT00000072630
|
Olr406
|
olfactory receptor 406 |
| chr7_+_35125424 | 2.19 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr8_-_16486419 | 2.17 |
ENSRNOT00000033223
|
LOC690235
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase-like 1) |
| chr19_-_59894937 | 2.16 |
ENSRNOT00000027145
|
Rbm34
|
RNA binding motif protein 34 |
| chr4_+_32541706 | 2.13 |
ENSRNOT00000050827
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmga2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 4.6 | 13.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 3.8 | 15.2 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 3.8 | 15.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 3.3 | 16.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 3.2 | 12.7 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 3.1 | 62.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 3.0 | 8.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 2.9 | 11.8 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.6 | 10.4 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 2.6 | 13.0 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 2.6 | 69.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 2.4 | 14.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 2.2 | 17.9 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 2.2 | 8.8 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 2.0 | 10.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 2.0 | 12.3 | GO:0099526 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 2.0 | 8.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.7 | 6.8 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.6 | 9.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.6 | 4.8 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.4 | 4.3 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 1.4 | 12.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 1.3 | 9.4 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) skeletal muscle satellite cell differentiation(GO:0014816) recognition of apoptotic cell(GO:0043654) |
| 1.2 | 3.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.1 | 20.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 1.1 | 6.7 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.0 | 4.9 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.9 | 3.5 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.9 | 10.5 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.8 | 7.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.8 | 19.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.8 | 9.0 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.7 | 8.7 | GO:0007135 | meiosis II(GO:0007135) |
| 0.7 | 2.8 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.7 | 8.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.7 | 4.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.6 | 5.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.6 | 2.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.6 | 7.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.6 | 1.8 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.6 | 7.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 6.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 2.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.5 | 2.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.5 | 2.9 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.5 | 5.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.4 | 2.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.4 | 1.3 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 | 2.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.4 | 7.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.4 | 18.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 60.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 3.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 2.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 1.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 3.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 3.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.3 | 8.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 7.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 4.9 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 2.8 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 1.4 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 2.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 1.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 0.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 2.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 1.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 3.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.2 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 9.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 10.7 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 3.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 16.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 4.7 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.1 | 10.7 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 2.8 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 2.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 2.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 3.8 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 3.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 6.1 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.1 | 1.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 12.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 3.5 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 5.6 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 3.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 8.8 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 2.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 2.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.7 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 3.2 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 2.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.0 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 4.1 | 12.3 | GO:0044317 | rod spherule(GO:0044317) |
| 3.5 | 62.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.8 | 16.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 1.2 | 15.1 | GO:0042627 | chylomicron(GO:0042627) |
| 1.1 | 13.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.1 | 4.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.0 | 15.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.9 | 18.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.8 | 4.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.8 | 3.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.7 | 4.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.7 | 13.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 10.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 1.8 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.5 | 2.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 12.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 9.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 1.8 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 3.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.4 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.4 | 11.8 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 2.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 2.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.4 | 9.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 2.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.4 | 12.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 14.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 9.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 2.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 1.5 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 2.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 4.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 8.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 1.8 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 8.7 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 8.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 4.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 6.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 3.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 7.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 4.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 14.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.3 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 2.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 17.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 10.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 8.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 8.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 1.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 5.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 6.1 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.1 | 3.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 7.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 12.6 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 21.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 17.4 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 4.2 | 62.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 3.6 | 10.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 2.7 | 16.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 2.5 | 7.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 2.4 | 7.2 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 2.3 | 6.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 2.2 | 8.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.1 | 10.4 | GO:0005534 | galactose binding(GO:0005534) |
| 2.0 | 59.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.7 | 6.7 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 1.6 | 23.0 | GO:0015250 | water channel activity(GO:0015250) |
| 1.4 | 4.3 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 1.3 | 9.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 5.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.2 | 9.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.2 | 3.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.0 | 3.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 1.0 | 6.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.0 | 8.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 2.6 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.9 | 4.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.8 | 2.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.8 | 2.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.8 | 5.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.8 | 8.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.8 | 10.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.8 | 12.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.8 | 16.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.8 | 23.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 4.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 9.5 | GO:0070097 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.7 | 14.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.7 | 4.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 12.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 1.8 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.6 | 1.8 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.6 | 9.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 13.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.6 | 12.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.6 | 3.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.5 | 23.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.5 | 4.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.5 | 3.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.5 | 10.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.4 | 1.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.4 | 4.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 2.8 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 5.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 2.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 4.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 3.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 18.2 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 7.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 5.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 17.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 2.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 51.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 11.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 10.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 25.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 8.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 2.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 4.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 8.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 8.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 16.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 8.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 5.6 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 59.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 2.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 8.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 12.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.6 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 65.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 5.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 10.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 6.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 6.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 45.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.1 | 12.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 10.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.9 | 16.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.8 | 31.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 19.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 9.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.6 | 7.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 2.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 4.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 10.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 9.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 6.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 3.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 12.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 13.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 5.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 14.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 4.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 7.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 10.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 3.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 4.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 5.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |