Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hmga1
Z-value: 1.01
Transcription factors associated with Hmga1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmga1
|
ENSRNOG00000000488 | high mobility group AT-hook 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga1 | rn6_v1_chr20_+_7136007_7136007 | 0.31 | 1.1e-08 | Click! |
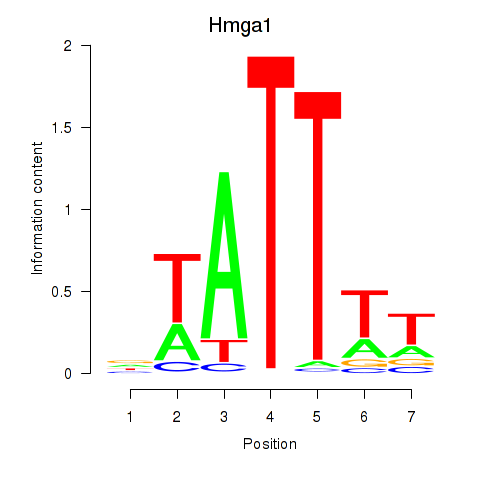
Activity profile of Hmga1 motif
Sorted Z-values of Hmga1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_28967980 | 46.75 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr15_+_17834635 | 40.66 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr6_+_107603580 | 39.48 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr4_-_135069970 | 37.32 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr16_-_73827488 | 33.31 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr2_-_231648122 | 30.82 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr11_+_20474483 | 26.98 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr8_-_53899669 | 26.56 |
ENSRNOT00000082257
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr4_-_16654811 | 26.23 |
ENSRNOT00000008637
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chrX_-_29648359 | 26.01 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr18_+_30562178 | 22.22 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr11_-_11213821 | 21.88 |
ENSRNOT00000093706
|
Robo2
|
roundabout guidance receptor 2 |
| chrX_+_84064427 | 21.34 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr11_-_4332255 | 21.03 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr8_-_8524643 | 20.78 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr19_-_11669578 | 20.59 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr9_-_28732919 | 19.93 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr14_-_3288017 | 19.70 |
ENSRNOT00000080452
|
LOC689986
|
hypothetical protein LOC689986 |
| chr6_+_73553210 | 19.25 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr2_-_18531210 | 18.86 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chrX_+_20423401 | 18.41 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr3_+_98297554 | 18.40 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr18_+_30509393 | 18.24 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr18_+_30587872 | 18.15 |
ENSRNOT00000040016
|
Pcdhb21
|
protocadherin beta 21 |
| chr14_+_7171613 | 16.87 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr18_+_29987206 | 16.84 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr18_+_30004565 | 16.62 |
ENSRNOT00000027393
|
Pcdha4
|
protocadherin alpha 4 |
| chrX_-_95062132 | 16.28 |
ENSRNOT00000049956
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr18_+_30574627 | 16.24 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr13_+_57243877 | 15.41 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chrX_+_53053609 | 15.40 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr3_-_66885085 | 15.11 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr18_+_13386133 | 15.05 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chr2_-_88763733 | 14.93 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr7_-_93280009 | 14.70 |
ENSRNOT00000064877
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr20_-_10013190 | 14.63 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chrX_-_142248369 | 14.22 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr19_-_14945302 | 14.19 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr10_+_97771264 | 14.09 |
ENSRNOT00000005257
|
Arsg
|
arylsulfatase G |
| chr12_-_5773036 | 14.07 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr1_-_255557055 | 13.85 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr5_-_128333805 | 13.84 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr5_+_2278357 | 13.68 |
ENSRNOT00000066590
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr6_-_61405195 | 13.04 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr13_-_90710148 | 12.95 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr14_+_48768537 | 12.66 |
ENSRNOT00000082599
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_-_10031167 | 12.63 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr13_+_98311827 | 12.06 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr9_-_115382445 | 11.90 |
ENSRNOT00000091316
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr10_-_18574909 | 11.83 |
ENSRNOT00000083090
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr11_-_31180616 | 11.81 |
ENSRNOT00000065535
ENSRNOT00000052033 ENSRNOT00000002812 |
Synj1
|
synaptojanin 1 |
| chr9_-_44419998 | 11.76 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr1_-_7443863 | 11.74 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr14_+_70780623 | 11.48 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr7_+_44009069 | 11.37 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr8_-_90962470 | 11.21 |
ENSRNOT00000086426
|
Lca5
|
LCA5, lebercilin |
| chr3_+_103753238 | 10.86 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr15_-_19541815 | 10.80 |
ENSRNOT00000008372
|
Txndc16
|
thioredoxin domain containing 16 |
| chr2_-_205212681 | 10.78 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr6_+_106496992 | 10.51 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr6_-_146470456 | 10.38 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr1_-_211923929 | 10.26 |
ENSRNOT00000054887
|
Nkx6-2
|
NK6 homeobox 2 |
| chr18_-_58423196 | 10.26 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr18_+_30864216 | 10.23 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr4_+_118655728 | 10.20 |
ENSRNOT00000043082
|
Aak1
|
AP2 associated kinase 1 |
| chr4_+_138269142 | 10.00 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr4_+_168832910 | 9.71 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr1_-_67134827 | 9.56 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr2_-_149444548 | 9.37 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr2_-_88553086 | 9.37 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr20_+_1764794 | 9.35 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr2_+_186980992 | 9.25 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr6_+_95323579 | 9.22 |
ENSRNOT00000007369
|
Pcnx4
|
pecanex homolog 4 (Drosophila) |
| chr11_+_45751812 | 8.76 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr4_-_45106267 | 8.51 |
ENSRNOT00000017099
|
AC123138.1
|
|
| chr10_+_99388130 | 8.37 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr18_-_26211445 | 8.26 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chrX_+_85338928 | 8.12 |
ENSRNOT00000074811
ENSRNOT00000006907 |
Dach2
|
dachshund family transcription factor 2 |
| chr2_+_193627243 | 8.08 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chr2_+_196013799 | 8.05 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr1_+_13261876 | 8.00 |
ENSRNOT00000090703
|
Reps1
|
RALBP1 associated Eps domain containing 1 |
| chr12_+_13097269 | 7.98 |
ENSRNOT00000087094
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr9_+_88493593 | 7.91 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr2_-_184263564 | 7.85 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr2_-_154418920 | 7.81 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr13_+_98231326 | 7.80 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr8_-_90984224 | 7.28 |
ENSRNOT00000044931
|
Lca5
|
LCA5, lebercilin |
| chr19_-_49510901 | 6.80 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr2_-_173800761 | 6.76 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chr4_+_24222500 | 6.62 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr8_+_58431407 | 6.47 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr1_+_257724842 | 6.45 |
ENSRNOT00000075515
|
AABR07006774.1
|
|
| chr8_-_126390801 | 5.80 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chrX_-_76708878 | 5.71 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr14_+_48726045 | 5.70 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_+_150135821 | 5.70 |
ENSRNOT00000047286
|
Zfp341
|
zinc finger protein 341 |
| chrX_+_54062935 | 5.55 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr3_-_48535909 | 5.52 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr2_+_186980793 | 5.46 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr9_-_65442257 | 5.44 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chrX_+_908044 | 5.38 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr7_+_74350479 | 5.35 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr1_-_128287151 | 5.19 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr16_-_3139177 | 5.17 |
ENSRNOT00000039311
|
Ccdc66
|
coiled-coil domain containing 66 |
| chr1_-_99135977 | 5.17 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chrX_+_33443186 | 5.08 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr3_+_65816569 | 5.08 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_-_67094567 | 5.00 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr16_-_6404578 | 4.97 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr3_+_154910626 | 4.97 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chr19_-_44078320 | 4.84 |
ENSRNOT00000026249
ENSRNOT00000089873 |
Cfdp1
|
craniofacial development protein 1 |
| chr17_+_2690062 | 4.60 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr6_-_137733026 | 4.55 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr14_-_81399353 | 4.45 |
ENSRNOT00000018340
|
Add1
|
adducin 1 |
| chr7_+_132378273 | 4.29 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr9_-_88816898 | 4.09 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr1_-_168611670 | 4.03 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr11_+_31389514 | 3.75 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr3_-_74371385 | 3.59 |
ENSRNOT00000041668
|
Olr518
|
olfactory receptor 518 |
| chr8_+_107508351 | 3.48 |
ENSRNOT00000081594
|
Cep70
|
centrosomal protein 70 |
| chr3_-_133122691 | 3.46 |
ENSRNOT00000083637
|
Tasp1
|
taspase 1 |
| chr1_-_168819946 | 3.36 |
ENSRNOT00000048499
|
Olr121
|
olfactory receptor 121 |
| chr3_-_76708430 | 3.31 |
ENSRNOT00000007890
|
Olr630
|
olfactory receptor 630 |
| chr13_-_32427177 | 3.28 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr1_-_172552099 | 3.22 |
ENSRNOT00000090373
|
Olr257
|
olfactory receptor 257 |
| chr20_+_18833481 | 3.21 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr13_+_44615553 | 3.21 |
ENSRNOT00000082439
ENSRNOT00000005289 |
Rab3gap1
|
RAB3 GTPase activating protein catalytic subunit 1 |
| chr15_-_40542495 | 3.15 |
ENSRNOT00000063943
|
Nup58
|
nucleoporin 58 |
| chr3_+_76468294 | 3.02 |
ENSRNOT00000037779
|
Olr619
|
olfactory receptor 619 |
| chr7_+_37101391 | 2.98 |
ENSRNOT00000029764
|
Eea1
|
early endosome antigen 1 |
| chrX_+_78042859 | 2.94 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr13_-_82005741 | 2.88 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr17_+_32516679 | 2.83 |
ENSRNOT00000032707
|
Serpinb9d
|
serine (or cysteine) peptidase inhibitor, clade B, member 9d |
| chr3_-_147188929 | 2.82 |
ENSRNOT00000055455
|
Rad21l1
|
RAD21 cohesin complex component like 1 |
| chr14_+_108143869 | 2.70 |
ENSRNOT00000090003
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr8_+_19810959 | 2.60 |
ENSRNOT00000049712
|
Olr1156
|
olfactory receptor 1156 |
| chr16_-_2227258 | 2.53 |
ENSRNOT00000089377
ENSRNOT00000067464 |
Slmap
|
sarcolemma associated protein |
| chr3_+_75987796 | 2.43 |
ENSRNOT00000091187
|
Olr594
|
olfactory receptor 594 |
| chr4_+_31333970 | 2.37 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr2_-_22096949 | 2.31 |
ENSRNOT00000089325
|
Zfyve16
|
zinc finger FYVE-type containing 16 |
| chr3_-_74545241 | 2.22 |
ENSRNOT00000090836
|
Olr531
|
olfactory receptor 531 |
| chr14_+_72889038 | 2.18 |
ENSRNOT00000084261
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr5_-_8864578 | 2.14 |
ENSRNOT00000008480
|
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr9_+_121831716 | 2.14 |
ENSRNOT00000056250
ENSRNOT00000056251 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr8_-_18115456 | 1.96 |
ENSRNOT00000045458
|
Olr1119
|
olfactory receptor 1119 |
| chr9_-_66019065 | 1.78 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr16_-_6405117 | 1.76 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr8_+_41285450 | 1.61 |
ENSRNOT00000076562
|
Olr1222
|
olfactory receptor 1222 |
| chr1_-_155955173 | 1.60 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr2_+_30666475 | 1.58 |
ENSRNOT00000076061
|
Taf9
|
TATA-box binding protein associated factor 9 |
| chr2_-_258932200 | 1.56 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr3_-_73401074 | 1.53 |
ENSRNOT00000012848
|
Olr476
|
olfactory receptor 476 |
| chr3_+_172385672 | 1.47 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr7_+_9262982 | 1.47 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr5_+_8916633 | 1.45 |
ENSRNOT00000066395
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr17_-_15511972 | 1.43 |
ENSRNOT00000074041
|
Aspnl1
|
asporin-like 1 |
| chr17_-_15429051 | 1.41 |
ENSRNOT00000074667
|
Nol8
|
nucleolar protein 8 |
| chrM_+_3904 | 1.41 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_-_168710460 | 1.40 |
ENSRNOT00000047399
|
Olr114
|
olfactory receptor 114 |
| chr6_-_44363915 | 1.40 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr2_+_204427608 | 1.38 |
ENSRNOT00000083374
|
Nhlh2
|
nescient helix loop helix 2 |
| chr9_-_73799427 | 1.35 |
ENSRNOT00000067589
|
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr3_-_77525518 | 1.32 |
ENSRNOT00000049513
|
Olr663
|
olfactory receptor 663 |
| chr7_-_73130740 | 1.31 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr1_-_216663720 | 1.25 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr5_-_50138475 | 1.18 |
ENSRNOT00000011969
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr3_-_150960030 | 1.17 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chrX_-_158288109 | 1.17 |
ENSRNOT00000072527
|
LOC103690175
|
PHD finger protein 6 |
| chr17_+_6667564 | 1.10 |
ENSRNOT00000025916
|
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr6_+_101532518 | 1.10 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr3_-_20883265 | 1.10 |
ENSRNOT00000052047
|
Olr414
|
olfactory receptor 414 |
| chr8_-_18051128 | 1.07 |
ENSRNOT00000073085
|
LOC100911944
|
olfactory receptor 7G1-like |
| chr4_+_10295122 | 1.07 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr17_-_53713408 | 1.07 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr14_-_102293677 | 0.96 |
ENSRNOT00000008135
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr14_-_33543238 | 0.95 |
ENSRNOT00000066382
|
Srp72
|
signal recognition particle 72 |
| chr4_-_167089055 | 0.90 |
ENSRNOT00000050409
|
Tas2r113
|
taste receptor, type 2, member 113 |
| chr7_-_142063212 | 0.87 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr11_-_61698595 | 0.84 |
ENSRNOT00000093445
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr8_-_109576353 | 0.83 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr16_-_70705128 | 0.74 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr8_-_19333047 | 0.67 |
ENSRNOT00000048500
|
Olr1144
|
olfactory receptor 1144 |
| chr8_-_19586811 | 0.66 |
ENSRNOT00000091744
|
Olr1149
|
olfactory receptor 1149 |
| chr3_+_76842898 | 0.65 |
ENSRNOT00000085537
|
Olr636
|
olfactory receptor 636 |
| chr1_-_60281386 | 0.64 |
ENSRNOT00000092184
|
Vom1r10
|
vomeronasal 1 receptor 10 |
| chr19_+_57614628 | 0.63 |
ENSRNOT00000026617
|
Gnpat
|
glyceronephosphate O-acyltransferase |
| chr20_+_1164821 | 0.62 |
ENSRNOT00000052052
|
LOC103690308
|
olfactory receptor 14J1-like |
| chr14_-_5006594 | 0.61 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr11_+_43394618 | 0.51 |
ENSRNOT00000051112
|
Olr1543
|
olfactory receptor 1543 |
| chr5_+_36566783 | 0.49 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr16_-_6404957 | 0.48 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr3_-_102589260 | 0.40 |
ENSRNOT00000091642
|
AC121203.1
|
|
| chr8_-_53816447 | 0.38 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr7_+_80750725 | 0.37 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr7_-_3707226 | 0.37 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr7_-_9427506 | 0.33 |
ENSRNOT00000041565
|
Olr1068
|
olfactory receptor 1068 |
| chr1_-_169845487 | 0.32 |
ENSRNOT00000039989
|
Olr179
|
olfactory receptor 179 |
| chr14_+_34455934 | 0.32 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chr1_-_170763377 | 0.31 |
ENSRNOT00000041335
|
Olr209
|
olfactory receptor 209 |
| chrX_-_37705263 | 0.25 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmga1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 7.7 | 30.8 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 6.6 | 19.9 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 6.1 | 18.4 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 5.2 | 26.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 5.1 | 15.4 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 4.8 | 19.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 4.4 | 21.9 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 4.4 | 26.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 3.6 | 10.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 3.5 | 14.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.4 | 13.7 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 3.1 | 9.4 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 2.6 | 7.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 2.3 | 46.8 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 2.1 | 10.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 2.0 | 14.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 2.0 | 13.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 2.0 | 11.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.9 | 5.7 | GO:1901580 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.8 | 5.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.7 | 5.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.7 | 5.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 1.6 | 8.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.4 | 19.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 1.4 | 4.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 1.3 | 5.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.2 | 3.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.2 | 15.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.1 | 3.2 | GO:1903233 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 1.1 | 34.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 1.0 | 27.0 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 1.0 | 144.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 1.0 | 2.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.9 | 6.5 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.9 | 4.4 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.9 | 9.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.9 | 7.9 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.8 | 7.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.8 | 13.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.7 | 2.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.7 | 21.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.7 | 8.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.6 | 24.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.6 | 10.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.6 | 11.8 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.6 | 4.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.5 | 11.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.5 | 2.5 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 1.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 20.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.5 | 4.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.5 | 18.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.5 | 1.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.4 | 27.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.4 | 1.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 2.8 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.3 | 1.8 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.3 | 1.1 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.3 | 0.8 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.2 | 14.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.2 | 1.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 14.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 10.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.2 | 33.8 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 10.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 3.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 14.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.0 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 2.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 2.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 16.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 10.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 14.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 14.7 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.1 | 2.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 2.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 9.6 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.1 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 0.2 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 3.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 11.8 | GO:0030031 | cell projection assembly(GO:0030031) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 7.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.2 | GO:0044317 | rod spherule(GO:0044317) |
| 6.4 | 19.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 3.3 | 33.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 3.3 | 19.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.9 | 5.7 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.6 | 7.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.2 | 46.2 | GO:0043034 | costamere(GO:0043034) |
| 1.0 | 10.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.0 | 14.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.8 | 37.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.8 | 5.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 2.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.7 | 13.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.7 | 13.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 21.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 11.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.6 | 7.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 8.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.6 | 21.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.5 | 22.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 30.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.5 | 3.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 45.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 4.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 2.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 9.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 144.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.3 | 1.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 6.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 16.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 13.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 8.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 18.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 10.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.5 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 7.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 26.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 115.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 15.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 11.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 4.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 12.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 3.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 11.5 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 11.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 6.1 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 3.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 33.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 3.9 | 11.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 3.8 | 15.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 3.7 | 22.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 2.8 | 11.0 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 2.6 | 7.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 2.6 | 12.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 2.1 | 19.2 | GO:0043495 | protein anchor(GO:0043495) |
| 2.0 | 14.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.8 | 21.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.6 | 7.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.5 | 18.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.5 | 26.2 | GO:0005522 | profilin binding(GO:0005522) |
| 1.4 | 11.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.4 | 4.1 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 1.3 | 9.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.2 | 7.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.2 | 26.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 1.1 | 5.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.1 | 10.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.0 | 10.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 1.0 | 18.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 1.0 | 2.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.9 | 18.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.8 | 5.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 15.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.8 | 10.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.8 | 35.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.8 | 19.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.7 | 2.9 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 5.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 15.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.5 | 8.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.5 | 13.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.4 | 18.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.4 | 11.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 14.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 24.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.3 | 13.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 0.9 | GO:0015099 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.3 | 1.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.3 | 11.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 27.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 4.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 16.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 130.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 0.9 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 5.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 9.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 19.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 21.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 5.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.2 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 4.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 8.0 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 20.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.8 | 18.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.6 | 20.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 11.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 26.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 14.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.4 | 18.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 11.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 9.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 22.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 10.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 16.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 8.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 18.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.4 | 19.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.4 | 14.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 1.1 | 10.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.0 | 12.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 1.0 | 33.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 34.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.8 | 21.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.7 | 35.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.7 | 21.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.7 | 11.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.5 | 13.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.5 | 14.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.5 | 4.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 5.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 21.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 18.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 11.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.3 | 5.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 7.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 4.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 16.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 11.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 3.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |