Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hmbox1
Z-value: 0.73
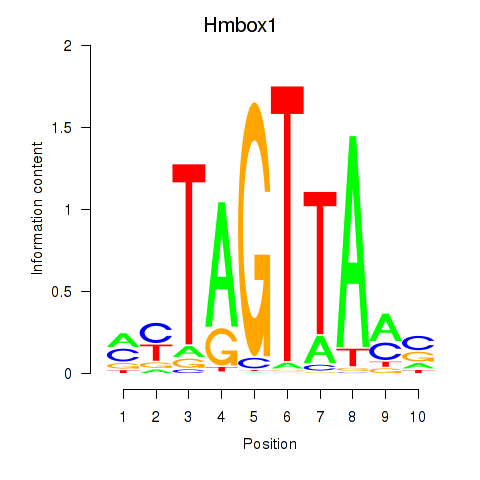
Transcription factors associated with Hmbox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmbox1
|
ENSRNOG00000013326 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmbox1 | rn6_v1_chr15_-_48284548_48284548 | 0.27 | 7.0e-07 | Click! |
Activity profile of Hmbox1 motif
Sorted Z-values of Hmbox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_139780386 | 65.52 |
ENSRNOT00000081147
|
AABR07041779.2
|
|
| chr17_-_69460321 | 27.04 |
ENSRNOT00000058367
|
Akr1c1
|
aldo-keto reductase family 1, member C1 |
| chr7_-_116425003 | 23.15 |
ENSRNOT00000076694
|
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr4_-_51199570 | 20.00 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr1_+_219964429 | 16.25 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr10_+_84871680 | 14.97 |
ENSRNOT00000073292
|
Prr15l
|
proline rich 15-like |
| chr2_+_114413410 | 13.25 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr1_-_13915594 | 11.93 |
ENSRNOT00000015927
|
Arfgef3
|
ARFGEF family member 3 |
| chr10_-_65692016 | 11.70 |
ENSRNOT00000085074
ENSRNOT00000038690 |
Slc13a2
|
solute carrier family 13 member 2 |
| chr20_+_20236151 | 10.38 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr1_+_23906717 | 9.83 |
ENSRNOT00000022798
|
Tcf21
|
transcription factor 21 |
| chr2_-_233743866 | 9.49 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr2_-_172459165 | 9.17 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr8_+_54925607 | 8.25 |
ENSRNOT00000059210
|
Plet1
|
placenta expressed transcript 1 |
| chr10_-_82852660 | 7.91 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chr8_+_107882219 | 7.53 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr10_+_90929423 | 7.17 |
ENSRNOT00000093180
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr9_-_27452902 | 7.14 |
ENSRNOT00000018325
|
Gsta1
|
glutathione S-transferase alpha 1 |
| chr20_-_44220702 | 6.94 |
ENSRNOT00000036853
|
Fam229b
|
family with sequence similarity 229, member B |
| chr18_+_32336102 | 6.81 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr18_+_70739492 | 6.75 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr1_+_127802978 | 6.73 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr9_+_117538009 | 6.66 |
ENSRNOT00000022878
|
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr10_+_90930010 | 6.17 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr8_-_73122740 | 6.14 |
ENSRNOT00000012026
|
Tln2
|
talin 2 |
| chr7_-_29070928 | 6.06 |
ENSRNOT00000080939
|
Chpt1
|
choline phosphotransferase 1 |
| chr14_-_114484127 | 6.05 |
ENSRNOT00000056706
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr4_-_31730386 | 5.97 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chrX_-_111179152 | 5.56 |
ENSRNOT00000089115
|
Morc4
|
MORC family CW-type zinc finger 4 |
| chr5_+_172488708 | 5.36 |
ENSRNOT00000019930
|
Morn1
|
MORN repeat containing 1 |
| chr20_+_28078500 | 5.33 |
ENSRNOT00000030005
|
Ccdc138
|
coiled-coil domain containing 138 |
| chr18_+_61377051 | 5.21 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chrX_+_40460047 | 5.20 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr15_+_47470863 | 5.16 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr8_-_47404010 | 5.07 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr9_+_117538346 | 4.89 |
ENSRNOT00000022849
|
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr4_-_118578815 | 4.89 |
ENSRNOT00000080502
|
Anxa4
|
annexin A4 |
| chr1_+_204617852 | 4.88 |
ENSRNOT00000022971
|
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr6_-_105261549 | 4.86 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr5_+_34040258 | 4.69 |
ENSRNOT00000009758
|
Ggh
|
gamma-glutamyl hydrolase |
| chr9_+_117795132 | 4.67 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr14_-_19132208 | 4.62 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr14_+_107785029 | 4.46 |
ENSRNOT00000013097
ENSRNOT00000087091 |
Fam161a
|
family with sequence similarity 161, member A |
| chr2_-_120316600 | 4.29 |
ENSRNOT00000084521
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr8_-_71337746 | 4.22 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr6_+_26686205 | 4.19 |
ENSRNOT00000082623
|
Slc5a6
|
solute carrier family 5 member 6 |
| chr1_+_229039889 | 4.19 |
ENSRNOT00000054800
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr9_+_81821346 | 4.18 |
ENSRNOT00000022234
|
Plcd4
|
phospholipase C, delta 4 |
| chr1_-_215284548 | 4.11 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr10_-_40764185 | 4.01 |
ENSRNOT00000017486
|
Sparc
|
secreted protein acidic and cysteine rich |
| chr8_-_114010277 | 3.81 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr13_+_82072497 | 3.79 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr7_-_107223047 | 3.66 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr17_-_78499881 | 3.65 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr2_+_208749996 | 3.65 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr15_-_3410423 | 3.65 |
ENSRNOT00000080274
|
Adk
|
adenosine kinase |
| chr4_+_117780533 | 3.58 |
ENSRNOT00000021211
|
Tprkb
|
Tp53rk binding protein |
| chr6_-_15191660 | 3.52 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr7_+_94795214 | 3.49 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr6_+_26686033 | 3.47 |
ENSRNOT00000081167
|
Slc5a6
|
solute carrier family 5 member 6 |
| chr19_-_601469 | 3.44 |
ENSRNOT00000016462
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr17_-_15566332 | 3.43 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr14_+_70164650 | 3.14 |
ENSRNOT00000004385
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr1_-_82610350 | 3.12 |
ENSRNOT00000028177
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chrX_+_25737292 | 3.08 |
ENSRNOT00000004893
|
RGD1563947
|
similar to RIKEN cDNA 4933400A11 |
| chr18_+_30840868 | 3.06 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr1_-_46329682 | 2.89 |
ENSRNOT00000046311
|
Tmem242
|
transmembrane protein 242 |
| chr15_-_77736892 | 2.88 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr17_+_54181419 | 2.81 |
ENSRNOT00000023861
|
Kif5b
|
kinesin family member 5B |
| chr13_-_104080631 | 2.81 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr3_+_101010899 | 2.76 |
ENSRNOT00000073258
|
Lin7c
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr3_-_122813583 | 2.75 |
ENSRNOT00000009681
|
Idh3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr9_+_121831716 | 2.72 |
ENSRNOT00000056250
ENSRNOT00000056251 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr16_+_50016857 | 2.70 |
ENSRNOT00000035781
|
Tlr3
|
toll-like receptor 3 |
| chr1_-_24302298 | 2.67 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr20_+_4374807 | 2.57 |
ENSRNOT00000084557
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr6_+_92229686 | 2.56 |
ENSRNOT00000046085
|
Atl1
|
atlastin GTPase 1 |
| chr10_+_96661696 | 2.56 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr20_+_28920616 | 2.50 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chrX_+_106390759 | 2.43 |
ENSRNOT00000049375
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr1_-_263269762 | 2.39 |
ENSRNOT00000022309
|
Got1
|
glutamic-oxaloacetic transaminase 1 |
| chr1_-_215536770 | 2.38 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr4_+_67378188 | 2.33 |
ENSRNOT00000030892
|
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr16_-_28716885 | 2.33 |
ENSRNOT00000059750
|
Spock3
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 3 |
| chr5_+_118574801 | 2.30 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_-_60088406 | 2.26 |
ENSRNOT00000091192
|
Vom1r7
|
vomeronasal 1 receptor 7 |
| chr8_+_49077053 | 2.24 |
ENSRNOT00000087922
|
Ift46
|
intraflagellar transport 46 |
| chr3_+_111135021 | 2.23 |
ENSRNOT00000018970
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr4_-_181348799 | 2.17 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr10_+_70629250 | 2.09 |
ENSRNOT00000013687
|
Rasl10b
|
RAS-like, family 10, member B |
| chr1_+_86429262 | 2.04 |
ENSRNOT00000045789
|
Vom1r3
|
vomeronasal 1 receptor 3 |
| chr10_+_77762900 | 2.03 |
ENSRNOT00000003308
|
Mmd
|
monocyte to macrophage differentiation-associated |
| chrX_-_31799560 | 2.03 |
ENSRNOT00000044103
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr2_-_89498395 | 2.01 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr13_-_89594738 | 1.98 |
ENSRNOT00000004641
|
Tomm40l
|
translocase of outer mitochondrial membrane 40 like |
| chr5_+_166464252 | 1.87 |
ENSRNOT00000055562
|
Ctnnbip1
|
catenin, beta-interacting protein 1 |
| chr4_-_182600258 | 1.84 |
ENSRNOT00000067970
|
Ergic2
|
ERGIC and golgi 2 |
| chr10_+_59743544 | 1.84 |
ENSRNOT00000093497
ENSRNOT00000056460 |
Tax1bp3
|
Tax1 binding protein 3 |
| chr2_+_189582991 | 1.63 |
ENSRNOT00000022627
|
Rab13
|
RAB13, member RAS oncogene family |
| chr1_-_221251329 | 1.58 |
ENSRNOT00000031678
|
Tigd3
|
tigger transposable element derived 3 |
| chr5_+_104394119 | 1.55 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chr8_+_59769994 | 1.53 |
ENSRNOT00000020309
|
Fbxo22
|
F-box protein 22 |
| chrX_+_96667863 | 1.53 |
ENSRNOT00000042552
|
RGD1561151
|
similar to hypothetical protein 4932411N23 |
| chr2_-_120316357 | 1.51 |
ENSRNOT00000065469
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr11_-_72109964 | 1.46 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr2_-_165600748 | 1.45 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chrM_+_5323 | 1.43 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr10_-_52290657 | 1.40 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr4_+_166986350 | 1.36 |
ENSRNOT00000040828
|
Tas2r117
|
taste receptor, type 2, member 117 |
| chr4_-_165828814 | 1.32 |
ENSRNOT00000007481
|
Tas2r105
|
taste receptor, type 2, member 105 |
| chr3_+_75712483 | 1.30 |
ENSRNOT00000049264
|
Olr575
|
olfactory receptor 575 |
| chr8_-_23130877 | 1.26 |
ENSRNOT00000019140
|
Elof1
|
elongation factor 1 homolog |
| chr6_-_55001464 | 1.25 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr4_-_147657847 | 1.19 |
ENSRNOT00000013889
|
Tmem40
|
transmembrane protein 40 |
| chr15_+_27330558 | 1.18 |
ENSRNOT00000085155
|
LOC103693383
|
olfactory receptor 11G2-like |
| chr19_-_37303788 | 1.16 |
ENSRNOT00000084734
|
Fhod1
|
formin homology 2 domain containing 1 |
| chr13_-_43566680 | 1.15 |
ENSRNOT00000073921
|
LOC100909582
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A-like |
| chr4_-_129544429 | 1.11 |
ENSRNOT00000091507
ENSRNOT00000083461 ENSRNOT00000087215 |
Tmf1
|
TATA element modulatory factor 1 |
| chr5_+_126976456 | 1.10 |
ENSRNOT00000014275
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr2_-_227582895 | 1.08 |
ENSRNOT00000020574
|
Mettl14
|
methyltransferase like 14 |
| chr4_-_157347803 | 1.06 |
ENSRNOT00000020785
|
Usp5
|
ubiquitin specific peptidase 5 |
| chr20_+_1889652 | 1.05 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chr3_+_38559914 | 1.04 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr19_+_551708 | 1.03 |
ENSRNOT00000015811
|
Fam96b
|
family with sequence similarity 96, member B |
| chr1_+_266482858 | 1.02 |
ENSRNOT00000027223
|
As3mt
|
arsenite methyltransferase |
| chr6_+_106052212 | 0.97 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr16_+_29674793 | 0.96 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr1_-_169882454 | 0.95 |
ENSRNOT00000043339
|
Olr181
|
olfactory receptor 181 |
| chrX_-_25628272 | 0.93 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr1_-_53014466 | 0.92 |
ENSRNOT00000049831
|
Sft2d1
|
SFT2 domain containing 1 |
| chr2_-_242828629 | 0.92 |
ENSRNOT00000043369
|
LOC499715
|
LRRGT00095 |
| chr5_+_129448879 | 0.89 |
ENSRNOT00000011765
|
Faf1
|
Fas associated factor 1 |
| chrX_-_70461553 | 0.86 |
ENSRNOT00000076110
ENSRNOT00000003872 ENSRNOT00000076812 |
Pdzd11
|
PDZ domain containing 11 |
| chr2_+_250995517 | 0.84 |
ENSRNOT00000018905
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr1_+_170205591 | 0.83 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr5_-_172488822 | 0.78 |
ENSRNOT00000019620
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr1_-_71173216 | 0.75 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr8_+_42443442 | 0.74 |
ENSRNOT00000015986
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr3_-_73124644 | 0.73 |
ENSRNOT00000012711
|
Olr455
|
olfactory receptor 455 |
| chr7_-_117257402 | 0.63 |
ENSRNOT00000081021
|
Plec
|
plectin |
| chr1_+_103064285 | 0.62 |
ENSRNOT00000075356
|
Vom1r70
|
vomeronasal 1 receptor 70 |
| chr6_-_131648661 | 0.59 |
ENSRNOT00000057179
|
LOC100910833
|
histone-lysine N-methyltransferase setd3-like |
| chr20_+_1944801 | 0.59 |
ENSRNOT00000075060
|
Olr1750
|
olfactory receptor 1750 |
| chr2_+_150106292 | 0.57 |
ENSRNOT00000073078
|
LOC100910567
|
arylacetamide deacetylase-like 2-like |
| chr4_-_166869399 | 0.54 |
ENSRNOT00000007521
|
Tas2r121
|
taste receptor, type 2, member 121 |
| chr6_-_115513354 | 0.47 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr4_-_165774126 | 0.45 |
ENSRNOT00000007453
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr17_-_69711689 | 0.42 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chrX_+_112311251 | 0.41 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr1_+_67340809 | 0.40 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr9_+_88493593 | 0.40 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr8_-_84506328 | 0.39 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr4_+_71795768 | 0.33 |
ENSRNOT00000031921
|
Tas2r135
|
taste receptor, type 2, member 135 |
| chr10_-_41491554 | 0.26 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr1_+_168623150 | 0.22 |
ENSRNOT00000021282
|
Olr107
|
olfactory receptor 107 |
| chrY_-_1305026 | 0.19 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr3_-_73854363 | 0.17 |
ENSRNOT00000051169
|
Olr496
|
olfactory receptor 496 |
| chr13_-_78885464 | 0.17 |
ENSRNOT00000003828
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr10_-_65200109 | 0.12 |
ENSRNOT00000030501
|
Nufip2
|
NUFIP2, FMR1 interacting protein 2 |
| chr17_-_55300998 | 0.11 |
ENSRNOT00000064331
|
Svil
|
supervillin |
| chr1_-_173469699 | 0.08 |
ENSRNOT00000040345
|
Olr281
|
olfactory receptor 281 |
| chr16_+_12510827 | 0.05 |
ENSRNOT00000077763
|
AABR07024716.1
|
|
| chr10_-_59743315 | 0.04 |
ENSRNOT00000093646
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr12_+_19314251 | 0.01 |
ENSRNOT00000001827
|
Ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr7_+_120106901 | 0.00 |
ENSRNOT00000063876
|
Gga1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmbox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 27.0 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 3.3 | 16.7 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 3.3 | 9.8 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 2.6 | 10.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.0 | 6.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 2.0 | 7.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.9 | 11.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 1.9 | 9.5 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.7 | 5.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 1.5 | 6.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.4 | 6.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.4 | 16.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.4 | 6.8 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.3 | 3.8 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 1.2 | 3.6 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) AMP salvage(GO:0044209) adenosine biosynthetic process(GO:0046086) |
| 1.2 | 20.0 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.1 | 9.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.0 | 7.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.8 | 2.4 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.8 | 6.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 | 2.2 | GO:1903588 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.7 | 3.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 3.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 2.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.7 | 4.9 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 | 2.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.7 | 2.7 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.6 | 3.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 2.8 | GO:1905150 | stress granule disassembly(GO:0035617) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 7.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.5 | 1.6 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.5 | 4.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.5 | 11.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 2.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 2.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 | 9.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.4 | 3.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) tetrahydrobiopterin biosynthetic process(GO:0006729) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.4 | 4.9 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.4 | 1.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.4 | 1.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 3.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 1.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) regulation of growth rate(GO:0040009) |
| 0.3 | 1.9 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.3 | 2.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 2.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 3.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 8.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.2 | 4.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.2 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 2.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 4.7 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.2 | 1.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 1.8 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 4.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 2.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 4.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 2.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.5 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.6 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 1.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.7 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 2.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.0 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 2.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 2.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.4 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 2.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 10.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 3.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 10.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 3.4 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.6 | 16.3 | GO:0008091 | spectrin(GO:0008091) |
| 1.3 | 10.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.0 | 4.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.9 | 2.8 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.8 | 3.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.7 | 11.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 2.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.7 | 4.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.7 | 9.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 2.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.6 | 2.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 6.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 2.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 34.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 7.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 20.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 4.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 2.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.2 | 3.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 10.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 3.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 5.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 4.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 4.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.6 | GO:1903293 | phosphatase complex(GO:1903293) |
| 0.0 | 9.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 10.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 6.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 23.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 27.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 2.0 | 7.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.5 | 13.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.3 | 20.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 1.2 | 4.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.2 | 6.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.0 | 3.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 1.0 | 4.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 1.0 | 6.8 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 1.0 | 7.7 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.8 | 2.4 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.8 | 6.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.7 | 2.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.7 | 4.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.7 | 2.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 3.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 3.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.6 | 2.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.6 | 3.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.6 | 2.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 3.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 11.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 4.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.4 | 1.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 9.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 7.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.4 | 2.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 3.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 41.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 6.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 4.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 2.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 9.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 4.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 2.0 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 2.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 4.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 5.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 5.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 11.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 11.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 3.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 4.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 4.6 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 4.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 2.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 3.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 4.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 6.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 6.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 7.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 31.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 13.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.8 | 26.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 11.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 6.8 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.4 | 5.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 2.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 6.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 7.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 8.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 2.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 2.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 2.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |