Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hivep1
Z-value: 1.08
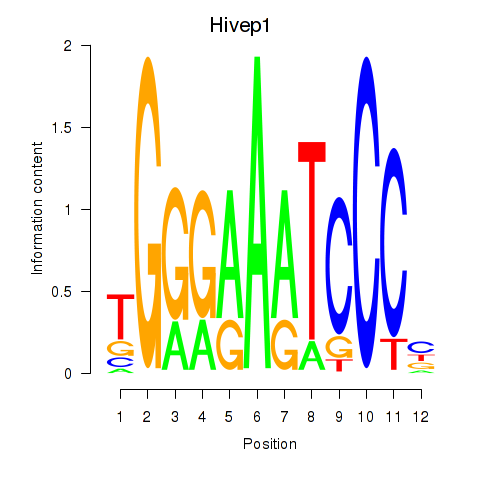
Transcription factors associated with Hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hivep1
|
ENSRNOG00000014460 | human immunodeficiency virus type I enhancer binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hivep1 | rn6_v1_chr17_-_22311392_22311392 | 0.10 | 8.1e-02 | Click! |
Activity profile of Hivep1 motif
Sorted Z-values of Hivep1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_163049084 | 77.23 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chr6_+_139405966 | 37.09 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr6_+_109562587 | 34.30 |
ENSRNOT00000011563
|
Batf
|
basic leucine zipper ATF-like transcription factor |
| chr20_-_4698718 | 34.28 |
ENSRNOT00000047527
|
RT1-CE7
|
RT1 class I, locus CE7 |
| chr20_-_4921348 | 32.74 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr20_+_2057878 | 31.88 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr4_+_69384145 | 31.32 |
ENSRNOT00000084834
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr20_-_157665 | 30.16 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr10_+_88764732 | 30.05 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr14_+_43810119 | 29.19 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_219097619 | 28.44 |
ENSRNOT00000078806
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr20_-_4863198 | 27.75 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr8_-_48850671 | 26.63 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr13_+_111870121 | 25.76 |
ENSRNOT00000007333
|
Irf6
|
interferon regulatory factor 6 |
| chr20_-_4896970 | 24.54 |
ENSRNOT00000061114
ENSRNOT00000060980 |
RT1-CE7
RT1-CE5
|
RT1 class I, locus CE7 RT1 class I, locus CE5 |
| chr20_-_4863011 | 24.19 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr20_+_5414448 | 23.81 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr8_-_77398156 | 23.30 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr4_-_115157263 | 23.13 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr20_+_2057689 | 22.89 |
ENSRNOT00000086007
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr17_+_56109549 | 21.84 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_+_63803309 | 21.69 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr10_+_39109522 | 21.34 |
ENSRNOT00000010968
|
Irf1
|
interferon regulatory factor 1 |
| chr1_-_80544825 | 21.18 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr6_-_76270457 | 20.97 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr20_-_3796258 | 20.92 |
ENSRNOT00000048076
ENSRNOT00000044520 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr6_-_140642221 | 20.88 |
ENSRNOT00000081996
|
AABR07065772.2
|
|
| chr1_+_252589785 | 20.82 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr20_+_5184515 | 20.64 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr18_-_11858744 | 20.53 |
ENSRNOT00000061417
ENSRNOT00000082891 |
Dsc2
|
desmocollin 2 |
| chr1_-_141471010 | 20.48 |
ENSRNOT00000082164
|
Plin1
|
perilipin 1 |
| chr5_+_168019058 | 20.03 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr5_-_150704117 | 19.72 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr1_+_199217016 | 18.73 |
ENSRNOT00000025732
|
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr10_-_83655182 | 18.27 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr20_+_2076267 | 18.09 |
ENSRNOT00000041500
|
RT1-M6-2
|
RT1 class I, locus M6, gene 2 |
| chr6_-_128727374 | 17.67 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr20_+_10438444 | 17.61 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr7_-_125795980 | 16.40 |
ENSRNOT00000017420
|
Arhgap8
|
Rho GTPase activating protein 8 |
| chr10_+_86860685 | 16.24 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr3_-_176958880 | 16.23 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr8_-_50531423 | 15.69 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr20_+_5509059 | 15.53 |
ENSRNOT00000065349
|
Kifc1
|
kinesin family member C1 |
| chr9_+_112360419 | 15.19 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chrX_+_54390733 | 15.12 |
ENSRNOT00000004977
|
RGD1565785
|
similar to chromosome X open reading frame 21 |
| chr3_+_95233874 | 13.46 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr4_-_23135354 | 13.33 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chrX_+_122724129 | 13.04 |
ENSRNOT00000017746
|
LOC100360218
|
interleukin 13 receptor, alpha 1-like |
| chr14_+_13192347 | 12.86 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr7_+_144078496 | 12.81 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr14_+_17210733 | 12.73 |
ENSRNOT00000003075
|
Cxcl10
|
C-X-C motif chemokine ligand 10 |
| chr1_-_166413564 | 12.54 |
ENSRNOT00000026286
|
Atg16l2
|
autophagy related 16-like 2 |
| chr6_-_138685656 | 12.52 |
ENSRNOT00000041706
|
AABR07065651.7
|
|
| chr1_-_261853495 | 11.07 |
ENSRNOT00000021144
|
Loxl4
|
lysyl oxidase-like 4 |
| chr4_+_153217782 | 10.95 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr15_+_34552410 | 10.79 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr1_+_164502389 | 10.56 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr18_+_80939875 | 10.22 |
ENSRNOT00000021729
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr5_-_107858104 | 9.19 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr3_+_138715570 | 8.99 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr5_-_56536772 | 8.82 |
ENSRNOT00000060765
|
Ddx58
|
DEXD/H-box helicase 58 |
| chr12_+_24978483 | 8.44 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr4_-_10995792 | 7.92 |
ENSRNOT00000078733
|
AABR07059243.1
|
|
| chr1_+_104635989 | 7.88 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr4_-_129846642 | 7.85 |
ENSRNOT00000010717
|
Frmd4b
|
FERM domain containing 4B |
| chr18_+_24397369 | 7.76 |
ENSRNOT00000022761
|
Sap130
|
Sin3A associated protein 130 |
| chr2_+_60169517 | 7.75 |
ENSRNOT00000080974
|
Prlr
|
prolactin receptor |
| chr7_+_14529482 | 7.64 |
ENSRNOT00000090844
ENSRNOT00000060284 ENSRNOT00000093239 |
Cyp4f5
Cyp4f17
|
cytochrome P450, family 4, subfamily f, polypeptide 5 cytochrome P450, family 4, subfamily f, polypeptide 17 |
| chr1_-_167347662 | 7.52 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr8_-_119265157 | 7.42 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr7_-_36499784 | 7.40 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr1_-_183630695 | 7.29 |
ENSRNOT00000036330
|
AABR07005506.1
|
|
| chr15_+_27739251 | 7.29 |
ENSRNOT00000011840
|
Parp2
|
poly (ADP-ribose) polymerase 2 |
| chr8_-_39093277 | 7.04 |
ENSRNOT00000043344
|
LOC100911068
|
roundabout homolog 4-like |
| chr10_-_65963932 | 6.98 |
ENSRNOT00000011726
|
Nlk
|
nemo like kinase |
| chr8_+_52753011 | 6.85 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr18_+_57011575 | 6.84 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr3_+_80676820 | 6.78 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr2_+_188253220 | 6.72 |
ENSRNOT00000027629
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr6_+_135866739 | 6.33 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr1_-_89590490 | 6.29 |
ENSRNOT00000028660
|
Gramd1a
|
GRAM domain containing 1A |
| chr16_-_50501921 | 6.18 |
ENSRNOT00000081023
|
Fat1
|
FAT atypical cadherin 1 |
| chr10_-_88611105 | 6.16 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr10_+_109446444 | 6.11 |
ENSRNOT00000066736
|
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chrX_+_111122552 | 6.00 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chrX_-_31138675 | 5.93 |
ENSRNOT00000004456
|
Fancb
|
Fanconi anemia, complementation group B |
| chr1_+_1784078 | 5.78 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr13_-_90977734 | 5.53 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr5_+_48011864 | 5.30 |
ENSRNOT00000067610
|
Mdn1
|
midasin AAA ATPase 1 |
| chr15_-_37056892 | 5.24 |
ENSRNOT00000047731
|
Zmym5
|
zinc finger MYM-type containing 5 |
| chr1_-_80716143 | 5.22 |
ENSRNOT00000092048
ENSRNOT00000025829 |
Cblc
|
Cbl proto-oncogene C |
| chr13_+_49870976 | 4.80 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr10_-_74769637 | 4.29 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chrX_-_38196060 | 4.06 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr14_-_18862407 | 4.05 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr2_+_188253378 | 3.99 |
ENSRNOT00000085690
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr3_-_29996865 | 3.78 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_72196437 | 3.75 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr18_-_24397551 | 3.61 |
ENSRNOT00000023032
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr3_+_47439076 | 3.58 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr20_-_4380084 | 3.48 |
ENSRNOT00000000499
|
Egfl8
|
EGF-like-domain, multiple 8 |
| chr6_-_47904437 | 3.46 |
ENSRNOT00000092867
|
Rps7
|
ribosomal protein S7 |
| chr1_-_86948845 | 3.37 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr1_-_178367828 | 3.35 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chr1_+_166433109 | 3.20 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr6_-_47904163 | 3.09 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr3_-_79121620 | 2.99 |
ENSRNOT00000087141
|
Olr741
|
olfactory receptor 741 |
| chr18_+_32273770 | 2.93 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr6_-_41039437 | 2.66 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr4_+_72637035 | 2.49 |
ENSRNOT00000007235
|
Olr819
|
olfactory receptor 819 |
| chr5_+_133864798 | 2.20 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr4_+_155654911 | 1.80 |
ENSRNOT00000087883
|
Foxj2
|
forkhead box J2 |
| chr9_-_4447715 | 1.77 |
ENSRNOT00000061882
|
Kat2b
|
lysine acetyltransferase 2B |
| chr10_-_88645364 | 1.75 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr18_-_32207749 | 1.72 |
ENSRNOT00000090882
ENSRNOT00000018843 |
Arhgap26
|
Rho GTPase activating protein 26 |
| chr17_+_88963981 | 1.69 |
ENSRNOT00000025053
|
Myo3a
|
myosin IIIA |
| chr5_+_98469047 | 1.65 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr1_-_252550394 | 1.48 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr18_-_36322320 | 1.47 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr15_-_3773340 | 1.34 |
ENSRNOT00000063831
|
LOC103689945
|
zinc finger SWIM domain-containing protein 8-like |
| chr12_-_38638536 | 1.28 |
ENSRNOT00000001690
|
Mlxip
|
MLX interacting protein |
| chr2_-_199563386 | 1.21 |
ENSRNOT00000075261
|
AABR07012616.1
|
|
| chr1_-_168994253 | 1.20 |
ENSRNOT00000039946
|
Hbg1
|
hemoglobin subunit gamma 1 |
| chr10_+_65448950 | 1.00 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr3_+_64190973 | 0.88 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr5_+_48012231 | 0.65 |
ENSRNOT00000077040
|
Mdn1
|
midasin AAA ATPase 1 |
| chr1_+_219764001 | 0.55 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr3_-_93216495 | 0.43 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr6_-_33507542 | 0.38 |
ENSRNOT00000007928
|
Gdf7
|
growth differentiation factor 7 |
| chr5_+_5866897 | 0.36 |
ENSRNOT00000011940
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hivep1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 32.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 7.9 | 23.8 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 7.8 | 23.3 | GO:0010034 | response to acetate(GO:0010034) |
| 7.3 | 29.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 7.2 | 21.7 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 6.7 | 20.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 6.0 | 42.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 5.8 | 23.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 5.5 | 77.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 5.2 | 15.7 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 4.9 | 19.7 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 4.2 | 12.7 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 4.2 | 21.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 4.2 | 20.8 | GO:0036337 | ovarian follicle atresia(GO:0001552) Fas signaling pathway(GO:0036337) |
| 4.1 | 16.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.6 | 10.7 | GO:1903699 | tarsal gland development(GO:1903699) |
| 3.6 | 160.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 3.3 | 13.0 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 3.1 | 34.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 2.9 | 17.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 2.8 | 28.4 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 2.8 | 8.4 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 2.6 | 10.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 2.6 | 20.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 2.4 | 7.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 2.3 | 16.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 2.2 | 17.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 2.2 | 8.8 | GO:0009597 | detection of virus(GO:0009597) |
| 2.1 | 12.5 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 2.0 | 7.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.9 | 5.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 1.7 | 13.3 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.5 | 6.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.5 | 21.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 1.5 | 5.9 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 1.4 | 12.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.4 | 26.6 | GO:0048535 | lymph node development(GO:0048535) |
| 1.4 | 33.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 1.4 | 5.5 | GO:0060266 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of neutrophil migration(GO:1902623) |
| 1.4 | 15.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 1.3 | 4.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 1.3 | 2.7 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 1.3 | 3.8 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.2 | 10.9 | GO:0001842 | neural fold formation(GO:0001842) |
| 1.2 | 3.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 1.2 | 3.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.8 | 18.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.7 | 2.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.7 | 6.6 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.6 | 1.8 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.6 | 20.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.5 | 9.2 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.5 | 6.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.5 | 3.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 6.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 6.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.4 | 6.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 3.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 | 15.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 1.5 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.4 | 7.4 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.3 | 7.6 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.3 | 10.2 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.3 | 7.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.3 | 4.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 | 12.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 7.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.3 | 1.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 6.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 6.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 3.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 7.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 9.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 16.4 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 7.0 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 11.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 10.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 4.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 22.6 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 1.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 5.2 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 28.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 7.0 | 111.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 5.1 | 45.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 4.2 | 20.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 2.6 | 15.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.6 | 23.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 2.5 | 17.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 2.5 | 19.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 2.4 | 21.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.7 | 8.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 10.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 1.0 | 23.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.9 | 20.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.8 | 10.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.8 | 6.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.6 | 9.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 7.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 12.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 149.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 7.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 5.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 22.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 6.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 3.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 15.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 11.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 4.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 7.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 14.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 21.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 6.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 10.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 4.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 7.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 23.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 102.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 6.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 13.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 72.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 5.8 | 23.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 5.2 | 15.7 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 4.1 | 24.5 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 3.9 | 23.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.5 | 10.6 | GO:0031893 | follicle-stimulating hormone receptor binding(GO:0031762) vasopressin receptor binding(GO:0031893) |
| 3.4 | 23.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 3.3 | 19.7 | GO:0070728 | leucine binding(GO:0070728) |
| 3.2 | 103.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 3.0 | 26.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 2.8 | 8.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 2.6 | 28.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 2.6 | 7.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.6 | 12.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.6 | 10.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) |
| 2.5 | 12.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.2 | 13.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.2 | 11.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 2.0 | 16.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.8 | 23.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 1.5 | 7.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.2 | 3.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.2 | 10.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 1.2 | 15.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 1.1 | 20.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 1.0 | 18.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.9 | 6.6 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.8 | 21.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.8 | 7.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.8 | 17.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 11.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 21.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.6 | 4.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 1.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.4 | 15.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 27.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 7.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 69.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 2.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 3.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 7.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 15.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 15.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 2.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 2.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 7.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 5.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 41.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 5.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 31.9 | GO:0000975 | regulatory region DNA binding(GO:0000975) |
| 0.1 | 7.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 6.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 5.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 10.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 9.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 5.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 2.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 3.7 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 24.1 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 6.0 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 41.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.8 | 30.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.7 | 21.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.4 | 15.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.1 | 19.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.8 | 23.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.7 | 21.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.7 | 20.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.7 | 21.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.6 | 18.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 16.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 7.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 7.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 7.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 3.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 45.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 4.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 9.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 7.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 11.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 39.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.0 | 30.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.8 | 33.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 1.5 | 20.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 1.4 | 20.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.3 | 75.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.9 | 21.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.8 | 41.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.7 | 15.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.5 | 15.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 7.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 10.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 17.7 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.3 | 9.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 7.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 5.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 16.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 4.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 6.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 1.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 18.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 6.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |