Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
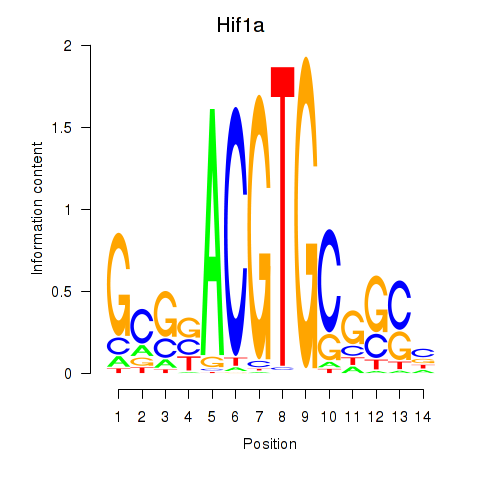
Results for Hif1a
Z-value: 0.67
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSRNOG00000008292 | hypoxia inducible factor 1 alpha subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | rn6_v1_chr6_+_96810907_96810907 | -0.44 | 1.2e-16 | Click! |
Activity profile of Hif1a motif
Sorted Z-values of Hif1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_58973020 | 21.01 |
ENSRNOT00000020379
|
Smtnl2
|
smoothelin-like 2 |
| chr4_-_115157263 | 18.68 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr19_+_25120128 | 16.67 |
ENSRNOT00000081243
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr5_-_24631679 | 16.45 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr4_-_10517832 | 14.42 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr1_+_33910912 | 14.31 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr1_+_102900286 | 13.94 |
ENSRNOT00000017468
|
Ldha
|
lactate dehydrogenase A |
| chr1_-_85220237 | 13.26 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr8_-_6203515 | 12.97 |
ENSRNOT00000087278
ENSRNOT00000031189 ENSRNOT00000008074 ENSRNOT00000085285 ENSRNOT00000007866 |
Yap1
|
yes-associated protein 1 |
| chr5_+_153507093 | 12.76 |
ENSRNOT00000086650
ENSRNOT00000083645 |
Runx3
|
runt-related transcription factor 3 |
| chr3_+_164822111 | 12.08 |
ENSRNOT00000014568
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr6_+_43829945 | 11.05 |
ENSRNOT00000086548
|
Klf11
|
Kruppel-like factor 11 |
| chr10_-_29450644 | 10.26 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr3_-_5976244 | 10.19 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr10_+_47961056 | 10.06 |
ENSRNOT00000027312
|
Fam83g
|
family with sequence similarity 83, member G |
| chr11_+_7210209 | 9.69 |
ENSRNOT00000076695
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr1_+_101397828 | 9.08 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr5_-_35831712 | 8.73 |
ENSRNOT00000010126
|
Prdm13
|
PR/SET domain 13 |
| chr3_-_5975734 | 8.58 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr9_+_19917603 | 8.25 |
ENSRNOT00000038239
|
Tdrd6
|
tudor domain containing 6 |
| chr8_+_116325286 | 8.03 |
ENSRNOT00000017461
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr10_+_46906115 | 7.79 |
ENSRNOT00000034006
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr3_+_119776925 | 7.52 |
ENSRNOT00000018549
|
Dusp2
|
dual specificity phosphatase 2 |
| chr5_+_169506138 | 7.40 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr4_-_132171153 | 7.34 |
ENSRNOT00000015058
ENSRNOT00000015075 |
Prok2
|
prokineticin 2 |
| chr2_-_84531192 | 7.10 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr4_+_140886545 | 7.08 |
ENSRNOT00000088273
|
Edem1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr3_+_6430201 | 6.94 |
ENSRNOT00000086352
|
Col5a1
|
collagen type V alpha 1 chain |
| chr4_+_120671489 | 6.91 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr7_+_144547939 | 6.59 |
ENSRNOT00000021580
|
Hoxc12
|
homeo box C12 |
| chr1_+_260732485 | 6.48 |
ENSRNOT00000076842
ENSRNOT00000076073 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr10_-_89338739 | 6.30 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr5_-_78379346 | 6.29 |
ENSRNOT00000020625
|
Alad
|
aminolevulinate dehydratase |
| chr7_-_75422268 | 6.24 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_263845762 | 5.77 |
ENSRNOT00000017268
|
Erlin1
|
ER lipid raft associated 1 |
| chr19_-_55367353 | 5.76 |
ENSRNOT00000091139
|
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr3_-_176958880 | 5.68 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr4_-_98593664 | 5.66 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr14_+_83510278 | 5.66 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr15_-_3435888 | 5.60 |
ENSRNOT00000016709
|
Adk
|
adenosine kinase |
| chr8_+_114897011 | 5.56 |
ENSRNOT00000074683
|
Twf2
|
twinfilin actin-binding protein 2 |
| chr6_+_128750795 | 5.53 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr3_+_147643250 | 5.47 |
ENSRNOT00000000013
|
Tcf15
|
transcription factor 15 |
| chr2_-_43393207 | 5.45 |
ENSRNOT00000017968
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr2_+_9526209 | 5.36 |
ENSRNOT00000021818
|
Lysmd3
|
LysM domain containing 3 |
| chr9_-_93404883 | 5.21 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr10_-_59049482 | 5.17 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr16_-_75855745 | 5.08 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr9_-_16612136 | 5.04 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr14_-_77321977 | 4.94 |
ENSRNOT00000084093
|
Lyar
|
Ly1 antibody reactive |
| chr12_-_31323810 | 4.90 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr14_-_80544342 | 4.82 |
ENSRNOT00000012304
|
Hmx1
|
H6 family homeobox 1 |
| chr11_+_80255790 | 4.82 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr6_+_52122085 | 4.70 |
ENSRNOT00000013043
|
Nampt
|
nicotinamide phosphoribosyltransferase |
| chr4_-_82127051 | 4.61 |
ENSRNOT00000083658
ENSRNOT00000007807 |
Hoxa1
|
homeo box A1 |
| chr19_+_39087990 | 4.57 |
ENSRNOT00000027553
|
Utp4
|
UTP4 small subunit processome component |
| chr6_+_134804141 | 4.56 |
ENSRNOT00000086143
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr13_-_90832469 | 4.47 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr14_-_77321783 | 4.43 |
ENSRNOT00000007154
|
Lyar
|
Ly1 antibody reactive |
| chr16_+_9563218 | 4.32 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr3_+_93495106 | 4.25 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr9_+_15068399 | 4.22 |
ENSRNOT00000064229
ENSRNOT00000087107 |
Foxp4
|
forkhead box P4 |
| chr10_-_83898527 | 3.99 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr2_-_148891900 | 3.71 |
ENSRNOT00000018488
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr13_-_90832138 | 3.69 |
ENSRNOT00000010930
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr17_+_36335804 | 3.67 |
ENSRNOT00000091334
|
E2f3
|
E2F transcription factor 3 |
| chr2_+_205783252 | 3.54 |
ENSRNOT00000025633
ENSRNOT00000025600 |
Trim33
|
tripartite motif-containing 33 |
| chr10_+_61432819 | 3.53 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr15_+_106257650 | 3.33 |
ENSRNOT00000014859
ENSRNOT00000072538 |
Ipo5
|
importin 5 |
| chr7_+_126939925 | 3.32 |
ENSRNOT00000022746
|
Gramd4
|
GRAM domain containing 4 |
| chr19_-_39087880 | 3.25 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr11_+_27004320 | 3.16 |
ENSRNOT00000002182
|
N6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr13_-_79088127 | 3.11 |
ENSRNOT00000034583
|
Prdx6
|
peroxiredoxin 6 |
| chr4_+_114807765 | 3.07 |
ENSRNOT00000084616
|
LOC103692169
|
39S ribosomal protein L53, mitochondrial |
| chr14_+_60657686 | 3.03 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr3_+_159305345 | 2.91 |
ENSRNOT00000008427
|
Srsf6
|
serine and arginine rich splicing factor 6 |
| chr8_+_55279373 | 2.87 |
ENSRNOT00000064290
|
Ppp2r1b
|
protein phosphatase 2 scaffold subunit A beta |
| chr20_+_46429222 | 2.72 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr9_+_16612433 | 2.68 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr10_+_59529785 | 2.43 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr10_+_72197977 | 2.40 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr9_-_16611899 | 2.39 |
ENSRNOT00000089168
|
Mea1
|
male-enhanced antigen 1 |
| chr1_-_129780356 | 2.36 |
ENSRNOT00000077479
|
Arrdc4
|
arrestin domain containing 4 |
| chr10_+_4411766 | 2.32 |
ENSRNOT00000003213
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr1_-_222178725 | 2.27 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr7_-_92882068 | 2.20 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr10_-_104054805 | 2.04 |
ENSRNOT00000004853
|
Nt5c
|
5', 3'-nucleotidase, cytosolic |
| chr3_+_151032952 | 2.01 |
ENSRNOT00000064013
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr2_-_44504354 | 2.01 |
ENSRNOT00000013035
|
Ddx4
|
DEAD-box helicase 4 |
| chr17_-_75886523 | 1.99 |
ENSRNOT00000046266
|
Usp6nl
|
USP6 N-terminal like |
| chr1_+_127802978 | 1.91 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr1_+_12823363 | 1.87 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr12_+_24669449 | 1.86 |
ENSRNOT00000044449
|
Wbscr22
|
Williams Beuren syndrome chromosome region 22 |
| chr9_-_61690956 | 1.79 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr10_-_35058870 | 1.79 |
ENSRNOT00000079481
|
AABR07029573.3
|
|
| chr4_-_129846642 | 1.75 |
ENSRNOT00000010717
|
Frmd4b
|
FERM domain containing 4B |
| chr3_+_9643047 | 1.73 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr8_+_22291445 | 1.70 |
ENSRNOT00000075174
ENSRNOT00000083194 |
Atg4d
|
autophagy related 4D, cysteine peptidase |
| chr8_+_44047592 | 1.60 |
ENSRNOT00000085084
|
Zfp202
|
zinc finger protein 202 |
| chr17_-_27112820 | 1.60 |
ENSRNOT00000018359
|
Bmp6
|
bone morphogenetic protein 6 |
| chr20_-_19825150 | 1.58 |
ENSRNOT00000032159
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr16_-_71125316 | 1.50 |
ENSRNOT00000020900
|
Plpp5
|
phospholipid phosphatase 5 |
| chr4_-_117234928 | 1.49 |
ENSRNOT00000073135
|
Pradc1
|
protease-associated domain containing 1 |
| chr3_+_6061940 | 1.43 |
ENSRNOT00000011556
|
Wdr5
|
WD repeat domain 5 |
| chr12_-_24669464 | 1.37 |
ENSRNOT00000043393
|
Dnajc30
|
DnaJ heat shock protein family (Hsp40) member C30 |
| chr1_+_163445527 | 1.33 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr1_+_219759183 | 1.32 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr14_+_61136746 | 1.22 |
ENSRNOT00000005126
|
Dhx15
|
DEAH-box helicase 15 |
| chr5_-_50362344 | 1.22 |
ENSRNOT00000035808
|
Zfp292
|
zinc finger protein 292 |
| chr7_-_24668250 | 1.16 |
ENSRNOT00000083806
|
Tmem263
|
transmembrane protein 263 |
| chr7_-_97759852 | 1.14 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr8_+_76977822 | 1.11 |
ENSRNOT00000090268
|
Sltm
|
SAFB-like, transcription modulator |
| chr10_+_39435227 | 0.87 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr5_-_136748980 | 0.70 |
ENSRNOT00000026844
|
Ipo13
|
importin 13 |
| chr7_-_11824742 | 0.58 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chrX_+_33599671 | 0.54 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr15_-_43667029 | 0.52 |
ENSRNOT00000014798
|
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr3_+_56125924 | 0.46 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_+_55687050 | 0.46 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr7_-_70467453 | 0.19 |
ENSRNOT00000052288
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr8_-_104221342 | 0.17 |
ENSRNOT00000015476
|
Atp1b3
|
ATPase Na+/K+ transporting subunit beta 3 |
| chr1_+_165170645 | 0.15 |
ENSRNOT00000022879
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr19_-_601469 | 0.14 |
ENSRNOT00000016462
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr9_+_16543688 | 0.12 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hif1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.7 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 3.6 | 14.3 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 3.4 | 10.3 | GO:0045819 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 2.4 | 7.1 | GO:1904380 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) endoplasmic reticulum mannose trimming(GO:1904380) |
| 2.3 | 13.9 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.9 | 7.8 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 1.9 | 5.6 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) AMP salvage(GO:0044209) adenosine biosynthetic process(GO:0046086) |
| 1.8 | 5.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.7 | 6.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.7 | 6.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.6 | 13.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.6 | 8.0 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.5 | 4.6 | GO:0021570 | rhombomere 3 development(GO:0021569) rhombomere 4 development(GO:0021570) |
| 1.4 | 14.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 1.4 | 5.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.4 | 5.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 1.2 | 4.8 | GO:0048294 | regulation of mast cell cytokine production(GO:0032763) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.0 | 6.3 | GO:0070541 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 1.0 | 6.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.0 | 5.8 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 1.0 | 5.8 | GO:0033634 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.9 | 1.8 | GO:0002424 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.7 | 11.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.7 | 2.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.7 | 4.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.7 | 5.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.7 | 2.0 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.6 | 4.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.6 | 2.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.6 | 12.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.6 | 1.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 0.6 | 1.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.5 | 16.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.5 | 3.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 2.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.5 | 2.9 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 18.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.5 | 3.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 5.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.5 | 1.9 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.5 | 2.7 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) response to water-immersion restraint stress(GO:1990785) |
| 0.4 | 3.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.4 | 5.5 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 4.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.4 | 3.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.4 | 1.6 | GO:2000860 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 4.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.3 | 2.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 7.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 7.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.2 | 3.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 7.3 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.2 | 9.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 3.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 4.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 5.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 1.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 13.3 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 | 6.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 4.3 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 2.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 2.3 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 4.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 10.7 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 11.9 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 8.2 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.7 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 0.0 | 3.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 21.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 5.1 | GO:0006639 | acylglycerol metabolic process(GO:0006639) |
| 0.0 | 0.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 9.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.2 | GO:0019532 | sulfate transport(GO:0008272) oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 2.1 | 18.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.7 | 6.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.4 | 5.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.9 | 4.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.9 | 7.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.8 | 13.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 2.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.5 | 10.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 8.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 3.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 6.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 5.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 4.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 1.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 6.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 8.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 4.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 13.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 10.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 9.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 12.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 7.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 14.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 43.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 5.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 17.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 7.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 10.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 6.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 5.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 7.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 3.1 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 3.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 3.5 | 13.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 3.4 | 10.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 3.2 | 9.7 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 2.7 | 8.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 2.1 | 6.3 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 1.9 | 7.8 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 1.2 | 4.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.2 | 4.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.9 | 5.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.8 | 2.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.6 | 7.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 5.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 1.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.6 | 5.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 6.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 2.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 6.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 7.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 1.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 14.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 5.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.4 | 13.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 18.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.4 | 4.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.4 | 6.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.4 | 3.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 5.6 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.4 | 1.8 | GO:0043559 | apolipoprotein A-I binding(GO:0034186) insulin binding(GO:0043559) |
| 0.3 | 3.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 8.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 0.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 3.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 5.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.0 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.2 | 10.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 1.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 4.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.2 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 2.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 5.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 3.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 20.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 5.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 9.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 11.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 7.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 6.5 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 23.4 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 4.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 2.7 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 4.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 13.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 4.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 4.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 16.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 10.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 6.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 6.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 10.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.6 | 8.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 13.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 7.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.5 | 6.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 6.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 12.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 5.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 6.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.3 | 6.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 4.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 10.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 9.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 6.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 10.7 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 2.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 5.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 5.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |