Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
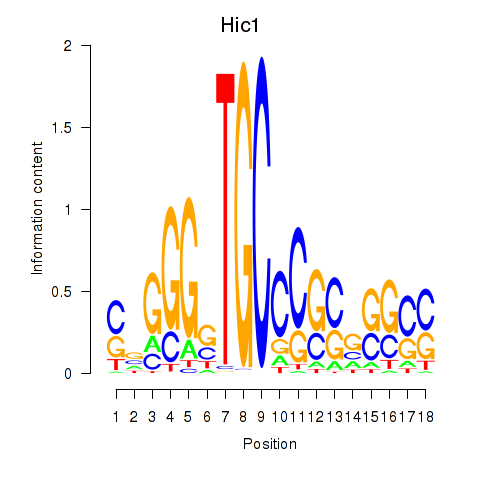
Results for Hic1
Z-value: 2.20
Transcription factors associated with Hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic1
|
ENSRNOG00000046405 | HIC ZBTB transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic1 | rn6_v1_chr10_-_62009582_62009582 | 0.05 | 3.4e-01 | Click! |
Activity profile of Hic1 motif
Sorted Z-values of Hic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_86297623 | 100.97 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr3_+_6773813 | 86.93 |
ENSRNOT00000065953
ENSRNOT00000013443 |
Olfm1
|
olfactomedin 1 |
| chr15_+_344360 | 85.55 |
ENSRNOT00000077671
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr4_+_99618622 | 73.88 |
ENSRNOT00000064772
|
Reep1
|
receptor accessory protein 1 |
| chr3_-_1924827 | 73.11 |
ENSRNOT00000006162
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr7_-_113937941 | 71.36 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr15_+_344685 | 68.97 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr6_-_105097054 | 65.79 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr1_+_226435979 | 60.79 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr2_-_24923128 | 59.57 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr2_-_229718659 | 56.01 |
ENSRNOT00000012676
|
Ugt8
|
UDP glycosyltransferase 8 |
| chr7_-_121029754 | 54.73 |
ENSRNOT00000004703
|
Nptxr
|
neuronal pentraxin receptor |
| chr4_+_9347528 | 52.87 |
ENSRNOT00000010521
|
Reln
|
reelin |
| chr4_+_58053041 | 52.84 |
ENSRNOT00000072698
|
Mest
|
mesoderm specific transcript |
| chr5_-_62621737 | 50.79 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr4_-_16130563 | 50.64 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr6_+_57516713 | 50.62 |
ENSRNOT00000063874
|
Dgkb
|
diacylglycerol kinase, beta |
| chr8_+_118333706 | 50.47 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr6_-_132958546 | 50.07 |
ENSRNOT00000041903
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr7_-_116607408 | 48.60 |
ENSRNOT00000076009
ENSRNOT00000056554 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr1_+_173607101 | 46.76 |
ENSRNOT00000074636
|
Tub
|
tubby bipartite transcription factor |
| chr5_+_64326733 | 46.66 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr14_-_78902063 | 45.99 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr1_+_1702696 | 45.72 |
ENSRNOT00000019181
|
Lrp11
|
LDL receptor related protein 11 |
| chr14_+_25589762 | 44.91 |
ENSRNOT00000043938
ENSRNOT00000067439 ENSRNOT00000002793 |
Epha5
|
EPH receptor A5 |
| chr9_-_85626094 | 44.74 |
ENSRNOT00000020919
|
Serpine2
|
serpin family E member 2 |
| chr14_+_60764409 | 43.51 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr5_+_156876706 | 42.96 |
ENSRNOT00000021864
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr7_+_128500011 | 42.85 |
ENSRNOT00000074625
|
Fam19a5
|
family with sequence similarity 19 member A5, C-C motif chemokine like |
| chr4_-_16130848 | 42.30 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chrX_-_106558366 | 41.78 |
ENSRNOT00000042126
|
Bex2
|
brain expressed X-linked 2 |
| chr4_-_145147397 | 41.75 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr1_-_220467159 | 41.71 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr5_-_166133274 | 41.61 |
ENSRNOT00000078830
|
Kif1b
|
kinesin family member 1B |
| chr8_-_117382715 | 41.14 |
ENSRNOT00000027466
|
P4htm
|
prolyl 4-hydroxylase, transmembrane |
| chr12_+_28381982 | 40.94 |
ENSRNOT00000076101
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr16_+_49266903 | 40.91 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr10_+_90342051 | 40.73 |
ENSRNOT00000028487
|
Rundc3a
|
RUN domain containing 3A |
| chr3_+_2262253 | 40.68 |
ENSRNOT00000042100
ENSRNOT00000061303 ENSRNOT00000048137 ENSRNOT00000048353 ENSRNOT00000012129 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr7_+_11556192 | 40.61 |
ENSRNOT00000046078
|
Diras1
|
DIRAS family GTPase 1 |
| chr5_-_17061361 | 40.51 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr1_-_31122093 | 40.49 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr3_+_145032200 | 40.44 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr1_-_261669584 | 40.31 |
ENSRNOT00000020568
ENSRNOT00000076555 |
Crtac1
|
cartilage acidic protein 1 |
| chr5_-_166133491 | 39.99 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr10_-_76813386 | 39.93 |
ENSRNOT00000032388
|
Nog
|
noggin |
| chr8_-_94564525 | 39.75 |
ENSRNOT00000084437
|
Snap91
|
synaptosomal-associated protein 91 |
| chr19_-_9777465 | 39.38 |
ENSRNOT00000017413
|
Ndrg4
|
NDRG family member 4 |
| chr19_+_58823814 | 39.23 |
ENSRNOT00000027058
ENSRNOT00000088626 |
Kcnk1
|
potassium two pore domain channel subfamily K member 1 |
| chr14_-_80732010 | 39.06 |
ENSRNOT00000012322
|
Adra2c
|
adrenoceptor alpha 2C |
| chr20_+_13044104 | 38.72 |
ENSRNOT00000066017
|
Dip2a
|
disco-interacting protein 2 homolog A |
| chr2_+_34186091 | 38.70 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr10_-_63952726 | 38.64 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr3_-_10602672 | 38.20 |
ENSRNOT00000011648
|
Ncs1
|
neuronal calcium sensor 1 |
| chr3_-_8924032 | 37.72 |
ENSRNOT00000023527
ENSRNOT00000085042 |
Sh3glb2
|
SH3 domain-containing GRB2-like endophilin B2 |
| chr7_-_136853957 | 37.68 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr3_+_142739781 | 37.59 |
ENSRNOT00000006181
|
Sstr4
|
somatostatin receptor 4 |
| chr1_-_112947162 | 37.39 |
ENSRNOT00000014573
ENSRNOT00000083894 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr16_-_64806050 | 36.69 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr7_-_76488216 | 36.66 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr17_-_18592750 | 36.52 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_-_55002322 | 36.12 |
ENSRNOT00000036092
|
Zdhhc2
|
zinc finger, DHHC-type containing 2 |
| chr11_+_30550141 | 36.08 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr4_+_147037179 | 35.85 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr1_-_200696928 | 35.79 |
ENSRNOT00000068183
ENSRNOT00000022253 ENSRNOT00000065448 ENSRNOT00000022331 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr1_-_64405149 | 35.68 |
ENSRNOT00000089944
|
Cacng7
|
calcium voltage-gated channel auxiliary subunit gamma 7 |
| chr10_-_110585376 | 35.68 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr2_-_172459165 | 35.62 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_-_5579894 | 35.53 |
ENSRNOT00000020044
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr9_+_70133969 | 35.43 |
ENSRNOT00000040525
|
Adam23
|
ADAM metallopeptidase domain 23 |
| chr8_-_94563760 | 34.94 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr5_+_166533181 | 34.78 |
ENSRNOT00000045063
|
Clstn1
|
calsyntenin 1 |
| chr9_+_82674202 | 34.44 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr10_-_68517564 | 34.30 |
ENSRNOT00000086961
|
Asic2
|
acid sensing ion channel subunit 2 |
| chr3_+_111597102 | 33.85 |
ENSRNOT00000081462
|
Tyro3
|
TYRO3 protein tyrosine kinase |
| chr17_+_13670520 | 33.66 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr15_-_42638392 | 33.55 |
ENSRNOT00000022020
|
Scara3
|
scavenger receptor class A, member 3 |
| chr3_-_2803574 | 33.17 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr10_+_11392625 | 32.97 |
ENSRNOT00000072802
|
Adcy9
|
adenylate cyclase 9 |
| chr8_+_65611570 | 32.97 |
ENSRNOT00000017147
|
Larp6
|
La ribonucleoprotein domain family, member 6 |
| chr5_-_58019836 | 32.81 |
ENSRNOT00000066977
|
Enho
|
energy homeostasis associated |
| chr17_+_41798783 | 32.44 |
ENSRNOT00000023519
|
Nrsn1
|
neurensin 1 |
| chr1_+_25173242 | 32.13 |
ENSRNOT00000016518
|
Clvs2
|
clavesin 2 |
| chr8_-_83421612 | 32.01 |
ENSRNOT00000015824
|
Hcrtr2
|
hypocretin receptor 2 |
| chr5_-_142857131 | 31.98 |
ENSRNOT00000038055
|
Maneal
|
mannosidase, endo-alpha-like |
| chr17_+_15924048 | 31.90 |
ENSRNOT00000050696
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr14_+_60857989 | 31.79 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr5_+_169519212 | 31.72 |
ENSRNOT00000024732
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr12_+_17416327 | 31.58 |
ENSRNOT00000089590
ENSRNOT00000092186 |
Adap1
|
ArfGAP with dual PH domains 1 |
| chr1_+_266953139 | 31.33 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr8_+_56179816 | 31.15 |
ENSRNOT00000059078
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr9_+_82647071 | 30.87 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr7_-_136853154 | 30.87 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr5_+_139783951 | 30.83 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr1_-_89560719 | 30.69 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr14_-_82287706 | 30.41 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr1_-_261575557 | 30.10 |
ENSRNOT00000076068
|
Crtac1
|
cartilage acidic protein 1 |
| chr14_+_87312203 | 30.06 |
ENSRNOT00000088032
|
Adcy1
|
adenylate cyclase 1 |
| chr10_+_70627401 | 30.04 |
ENSRNOT00000076897
|
Rasl10b
|
RAS-like, family 10, member B |
| chr15_+_37806836 | 30.03 |
ENSRNOT00000076285
|
Il17d
|
interleukin 17D |
| chr16_-_6405117 | 29.91 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr4_-_146016325 | 29.85 |
ENSRNOT00000067931
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr4_+_22445414 | 29.72 |
ENSRNOT00000087657
ENSRNOT00000030224 |
Rundc3b
|
RUN domain containing 3B |
| chr6_-_94980004 | 29.62 |
ENSRNOT00000006373
|
Rtn1
|
reticulon 1 |
| chr1_-_15301998 | 29.62 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr6_+_126018841 | 29.60 |
ENSRNOT00000089840
ENSRNOT00000088089 |
Slc24a4
|
solute carrier family 24 member 4 |
| chr4_+_66405166 | 29.46 |
ENSRNOT00000077486
ENSRNOT00000067097 |
Clec2l
|
C-type lectin domain family 2, member L |
| chr3_-_165477771 | 29.46 |
ENSRNOT00000071672
|
Atp9a
|
ATPase phospholipid transporting 9A (putative) |
| chr5_-_165083487 | 29.45 |
ENSRNOT00000036647
|
Disp3
|
dispatched RND transporter family member 3 |
| chr8_+_56179637 | 29.34 |
ENSRNOT00000035989
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr19_-_58399816 | 29.30 |
ENSRNOT00000026843
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr12_-_24710019 | 29.22 |
ENSRNOT00000049601
|
Stx1a
|
syntaxin 1A |
| chr13_-_73819896 | 28.92 |
ENSRNOT00000036392
|
Fam163a
|
family with sequence similarity 163, member A |
| chr9_-_45505767 | 28.80 |
ENSRNOT00000033964
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr1_+_113034227 | 28.75 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr3_-_147143576 | 28.66 |
ENSRNOT00000091811
ENSRNOT00000012727 |
Snph
|
syntaphilin |
| chr3_-_11885311 | 28.66 |
ENSRNOT00000021189
ENSRNOT00000021178 |
Stxbp1
|
syntaxin binding protein 1 |
| chr3_-_104502471 | 28.57 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr8_+_119566509 | 28.45 |
ENSRNOT00000028633
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr8_+_83070545 | 28.35 |
ENSRNOT00000014926
|
Hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chrX_+_20351486 | 28.29 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr19_-_11669578 | 28.21 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr1_-_212622537 | 27.98 |
ENSRNOT00000025609
|
Sprn
|
shadow of prion protein homolog (zebrafish) |
| chr2_-_186245342 | 27.95 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr12_+_22026075 | 27.84 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr8_+_79606789 | 27.74 |
ENSRNOT00000087114
|
Pygo1
|
pygopus family PHD finger 1 |
| chr5_-_172769421 | 27.66 |
ENSRNOT00000021285
|
Prkcz
|
protein kinase C, zeta |
| chr1_-_146556171 | 27.66 |
ENSRNOT00000017636
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr3_-_92290919 | 27.57 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr17_-_10208360 | 27.46 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr8_+_128972311 | 27.46 |
ENSRNOT00000025460
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr3_+_124896618 | 27.30 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr9_+_82120059 | 27.26 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr1_-_270472866 | 27.20 |
ENSRNOT00000015979
|
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chrX_+_106823491 | 27.05 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr1_+_263186235 | 26.98 |
ENSRNOT00000021876
|
Cnnm1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr3_+_79713567 | 26.94 |
ENSRNOT00000012110
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr16_-_20439206 | 26.89 |
ENSRNOT00000026391
|
Rab3a
|
RAB3A, member RAS oncogene family |
| chr4_+_9347779 | 26.87 |
ENSRNOT00000061858
|
Reln
|
reelin |
| chrX_-_10772633 | 26.73 |
ENSRNOT00000035153
|
Ube2q2l
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr9_-_94601852 | 26.59 |
ENSRNOT00000022485
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr12_+_943006 | 26.48 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr7_-_135630654 | 26.44 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_-_84491466 | 26.28 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr19_+_52086325 | 26.17 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr7_-_116607674 | 26.12 |
ENSRNOT00000076014
|
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr8_+_408001 | 26.11 |
ENSRNOT00000046058
|
Gucy1a2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr1_-_112947399 | 26.08 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr3_+_22640545 | 26.05 |
ENSRNOT00000064507
ENSRNOT00000014452 |
Lhx2
|
LIM homeobox 2 |
| chr3_+_161433410 | 25.96 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr15_-_62200837 | 25.93 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr12_-_12025549 | 25.73 |
ENSRNOT00000001331
|
Nptx2
|
neuronal pentraxin 2 |
| chr13_+_69797880 | 25.71 |
ENSRNOT00000030109
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr9_+_17841410 | 25.62 |
ENSRNOT00000031706
|
Tmem151b
|
transmembrane protein 151B |
| chr19_-_57192095 | 25.52 |
ENSRNOT00000058080
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr13_+_110920830 | 25.39 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr8_+_6811543 | 25.36 |
ENSRNOT00000008905
ENSRNOT00000042553 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr1_-_91462603 | 25.22 |
ENSRNOT00000015384
|
Lrp3
|
LDL receptor related protein 3 |
| chr7_+_11737293 | 25.10 |
ENSRNOT00000046059
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr10_+_39655455 | 25.07 |
ENSRNOT00000058817
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_42587666 | 24.96 |
ENSRNOT00000083225
ENSRNOT00000025355 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr10_-_108691367 | 24.94 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr1_+_224882439 | 24.91 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr18_-_18079560 | 24.86 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr15_+_19547871 | 24.71 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr2_-_179704629 | 24.61 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr6_-_122897997 | 24.52 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr5_-_39215102 | 24.34 |
ENSRNOT00000050653
|
Klhl32
|
kelch-like family member 32 |
| chr8_+_28352772 | 24.09 |
ENSRNOT00000012391
|
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr10_-_14937336 | 24.04 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr15_-_43542939 | 23.93 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr2_-_26699333 | 23.91 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr13_-_111765944 | 23.87 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr1_-_241875864 | 23.86 |
ENSRNOT00000091282
|
Fam189a2
|
family with sequence similarity 189, member A2 |
| chr6_-_65319527 | 23.82 |
ENSRNOT00000005618
|
Stxbp6
|
syntaxin binding protein 6 |
| chr18_-_63357194 | 23.64 |
ENSRNOT00000089408
ENSRNOT00000066103 |
Spire1
|
spire-type actin nucleation factor 1 |
| chr17_+_18059382 | 23.59 |
ENSRNOT00000031656
|
Nhlrc1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr3_-_124724252 | 23.48 |
ENSRNOT00000028885
|
Slc23a2
|
solute carrier family 23 member 2 |
| chr8_+_111600532 | 23.45 |
ENSRNOT00000081952
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr13_+_101181994 | 23.39 |
ENSRNOT00000052407
|
Susd4
|
sushi domain containing 4 |
| chr2_-_186245163 | 23.38 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr10_-_14788617 | 23.27 |
ENSRNOT00000043626
|
Cacna1h
|
calcium voltage-gated channel subunit alpha1 H |
| chr1_-_102106127 | 23.24 |
ENSRNOT00000028685
|
Kcnj11
|
potassium voltage-gated channel subfamily J member 11 |
| chr14_+_77380262 | 23.24 |
ENSRNOT00000008030
|
Nsg1
|
neuron specific gene family member 1 |
| chr5_-_58078545 | 23.17 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr1_+_162817611 | 23.04 |
ENSRNOT00000091952
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chrX_+_76083549 | 23.01 |
ENSRNOT00000003573
|
Magee1
|
MAGE family member E1 |
| chr6_+_133576568 | 22.98 |
ENSRNOT00000085933
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr1_-_112811936 | 22.92 |
ENSRNOT00000093339
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr4_+_146106386 | 22.87 |
ENSRNOT00000008342
|
Slc6a11
|
solute carrier family 6 member 11 |
| chr20_-_8574082 | 22.66 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr19_+_37305248 | 22.61 |
ENSRNOT00000068299
|
Slc9a5
|
solute carrier family 9 member A5 |
| chrX_+_40363646 | 22.55 |
ENSRNOT00000010135
|
Sms
|
spermine synthase |
| chr18_-_37819097 | 22.38 |
ENSRNOT00000025880
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr7_-_126465723 | 22.24 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr5_+_148193710 | 22.22 |
ENSRNOT00000088568
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr14_-_78308933 | 22.19 |
ENSRNOT00000065334
|
Crmp1
|
collapsin response mediator protein 1 |
| chr14_+_78640620 | 22.19 |
ENSRNOT00000034730
|
Wfs1
|
wolframin ER transmembrane glycoprotein |
| chr8_-_103190243 | 21.91 |
ENSRNOT00000075305
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr14_+_60123169 | 21.76 |
ENSRNOT00000006610
|
Sel1l3
|
SEL1L family member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 51.5 | 154.5 | GO:0060082 | eye blink reflex(GO:0060082) |
| 33.7 | 101.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 20.3 | 60.8 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 18.7 | 56.0 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 16.9 | 101.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 16.3 | 81.6 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 16.2 | 145.5 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 15.5 | 92.9 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 14.9 | 59.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 13.5 | 40.5 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 13.3 | 39.9 | GO:1905006 | regulation of cytokine activity(GO:0060300) fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 12.2 | 36.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 12.2 | 73.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 12.0 | 48.0 | GO:0021586 | pons maturation(GO:0021586) |
| 11.2 | 44.7 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 10.8 | 43.3 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 10.7 | 74.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 10.6 | 21.3 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 10.4 | 20.8 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 10.2 | 40.9 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 10.1 | 50.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 10.0 | 30.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 9.4 | 28.3 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 8.9 | 17.9 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 8.9 | 70.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 8.4 | 25.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 8.3 | 24.9 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 7.9 | 23.6 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 7.8 | 15.7 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 7.8 | 23.5 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 7.8 | 23.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 7.8 | 23.4 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 7.5 | 15.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 7.4 | 22.2 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 7.4 | 22.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 7.3 | 44.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 7.2 | 21.6 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 7.1 | 28.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 7.1 | 21.2 | GO:0061744 | motor behavior(GO:0061744) |
| 7.0 | 111.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 6.9 | 41.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 6.8 | 20.3 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 6.7 | 40.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 6.7 | 26.9 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 6.4 | 19.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 6.3 | 37.6 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 6.2 | 12.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 6.2 | 80.6 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 6.1 | 104.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 6.0 | 24.0 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 6.0 | 29.9 | GO:1901660 | calcium ion export(GO:1901660) |
| 5.9 | 23.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 5.9 | 5.9 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 5.8 | 17.4 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 5.8 | 23.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 5.6 | 11.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 5.6 | 33.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 5.6 | 39.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 5.6 | 33.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 5.6 | 11.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 5.5 | 22.2 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 5.5 | 27.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 5.5 | 21.9 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 5.4 | 10.9 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 5.2 | 20.7 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 5.1 | 40.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 5.0 | 24.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 4.9 | 34.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 4.7 | 28.5 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 4.6 | 27.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 4.6 | 32.0 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 4.6 | 13.7 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 4.4 | 17.6 | GO:0036394 | amylase secretion(GO:0036394) |
| 4.4 | 17.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 4.4 | 39.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 4.4 | 4.4 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 4.3 | 26.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 4.3 | 77.5 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) positive regulation of neurotransmitter transport(GO:0051590) |
| 4.3 | 21.5 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 4.3 | 12.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 4.2 | 12.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 4.2 | 12.7 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 4.1 | 20.7 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 4.1 | 73.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 4.1 | 20.5 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 4.1 | 172.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 4.0 | 23.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 4.0 | 102.8 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 3.9 | 15.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 3.9 | 54.9 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 3.9 | 7.8 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 3.9 | 38.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 3.9 | 38.6 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 3.9 | 15.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 3.8 | 22.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 3.8 | 3.8 | GO:0008355 | olfactory learning(GO:0008355) |
| 3.8 | 11.3 | GO:0061441 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) metanephric part of ureteric bud development(GO:0035502) renal artery morphogenesis(GO:0061441) kidney smooth muscle tissue development(GO:0072194) |
| 3.8 | 18.8 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 3.7 | 18.5 | GO:0046959 | habituation(GO:0046959) |
| 3.7 | 11.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 3.6 | 10.9 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 3.6 | 25.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 3.6 | 10.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 3.5 | 7.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 3.5 | 38.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 3.5 | 17.4 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 3.5 | 13.9 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 3.4 | 17.0 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) |
| 3.4 | 10.2 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 3.4 | 13.5 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 3.3 | 10.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 3.3 | 9.9 | GO:0032898 | neurotrophin production(GO:0032898) nerve growth factor production(GO:0032902) |
| 3.3 | 46.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 3.3 | 45.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 3.3 | 13.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 3.2 | 9.7 | GO:1990401 | regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling(GO:0060683) embryonic lung development(GO:1990401) |
| 3.2 | 9.7 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 3.2 | 44.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 3.2 | 44.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 3.2 | 6.4 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 3.1 | 18.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 3.0 | 21.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 3.0 | 8.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 3.0 | 8.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.9 | 14.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 2.9 | 5.8 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 2.8 | 8.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of osteoclast development(GO:2001205) |
| 2.8 | 50.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 2.8 | 8.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 2.8 | 19.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 2.8 | 27.6 | GO:0007614 | short-term memory(GO:0007614) |
| 2.7 | 21.6 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 2.6 | 18.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 2.6 | 15.8 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 2.6 | 7.9 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 2.6 | 44.5 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 2.6 | 26.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 2.6 | 12.9 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 2.6 | 30.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.5 | 10.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 2.5 | 12.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 2.5 | 15.1 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 2.5 | 10.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 2.5 | 5.0 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 2.5 | 12.5 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 2.5 | 22.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 2.4 | 24.3 | GO:0015886 | heme transport(GO:0015886) |
| 2.4 | 21.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 2.4 | 7.2 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 2.4 | 14.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.4 | 14.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 2.4 | 7.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.4 | 7.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 2.3 | 96.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 2.3 | 2.3 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 2.3 | 31.9 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 2.2 | 6.7 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 2.2 | 6.5 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 2.1 | 15.0 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 2.1 | 10.7 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 2.1 | 23.2 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 2.1 | 58.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 2.1 | 35.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 2.1 | 22.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 2.1 | 66.3 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 2.1 | 26.8 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 2.0 | 26.6 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 2.0 | 10.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 2.0 | 12.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 2.0 | 26.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 2.0 | 9.9 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 2.0 | 5.9 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) threonyl-tRNA aminoacylation(GO:0006435) |
| 1.9 | 9.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 1.9 | 3.8 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 1.9 | 7.6 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 1.9 | 22.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.8 | 7.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 1.8 | 55.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 1.8 | 5.5 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 1.8 | 35.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 1.7 | 6.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.7 | 20.7 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 1.7 | 13.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.7 | 5.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 1.7 | 5.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.7 | 35.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 1.7 | 5.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.6 | 9.8 | GO:0042747 | circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 1.6 | 9.8 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.6 | 4.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.6 | 43.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 1.6 | 17.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.5 | 1.5 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.5 | 12.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 1.5 | 13.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.5 | 10.6 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 1.5 | 4.5 | GO:0060214 | endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 1.5 | 17.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.5 | 5.9 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.4 | 15.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.4 | 2.9 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 1.4 | 21.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 1.4 | 29.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.4 | 5.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 1.4 | 29.5 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 1.4 | 18.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.4 | 24.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 1.3 | 23.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 1.3 | 31.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 1.3 | 39.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 1.3 | 16.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 1.3 | 2.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 1.3 | 25.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 1.3 | 2.5 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 1.2 | 3.7 | GO:0070428 | regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.2 | 6.2 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.2 | 15.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 1.2 | 3.6 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 1.2 | 13.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 1.2 | 30.2 | GO:0097009 | energy homeostasis(GO:0097009) |
| 1.1 | 19.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 1.1 | 4.5 | GO:0003383 | apical constriction(GO:0003383) |
| 1.1 | 4.5 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 1.1 | 7.8 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 1.1 | 10.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 1.1 | 5.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 1.1 | 4.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.1 | 6.4 | GO:0010045 | response to nickel cation(GO:0010045) |
| 1.0 | 3.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.0 | 22.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 1.0 | 11.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.0 | 26.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 1.0 | 3.0 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 1.0 | 6.9 | GO:0061042 | vascular wound healing(GO:0061042) |
| 1.0 | 16.8 | GO:0003159 | morphogenesis of an endothelium(GO:0003159) endothelial tube morphogenesis(GO:0061154) |
| 1.0 | 5.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.0 | 17.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.0 | 1.9 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.9 | 3.8 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.9 | 12.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.9 | 1.9 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.9 | 8.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.9 | 1.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.9 | 2.8 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.9 | 13.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.9 | 21.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.9 | 17.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.9 | 8.8 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.9 | 1.8 | GO:1900625 | monocyte aggregation(GO:0070487) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.9 | 3.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.9 | 6.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.9 | 8.6 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.9 | 2.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.9 | 3.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.8 | 18.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.8 | 6.7 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.8 | 33.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.8 | 21.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.8 | 11.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.8 | 4.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) |
| 0.8 | 2.5 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) |
| 0.8 | 13.8 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.8 | 1.6 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.8 | 25.0 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.8 | 33.7 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.8 | 9.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.8 | 29.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.8 | 5.5 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.8 | 6.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.8 | 18.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.8 | 4.6 | GO:0021684 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 43.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.8 | 5.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.8 | 3.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.8 | 10.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.7 | 5.9 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.7 | 3.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.7 | 2.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.7 | 2.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.7 | 10.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.7 | 15.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.7 | 2.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.7 | 6.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.7 | 5.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.7 | 12.5 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.7 | 5.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.7 | 13.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.7 | 6.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.7 | 6.0 | GO:0030002 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 | 12.9 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.6 | 80.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.6 | 3.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.6 | 29.7 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.6 | 8.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.6 | 6.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.6 | 3.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.6 | 20.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.6 | 8.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.6 | 4.7 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 31.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.6 | 2.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.6 | 5.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.6 | 2.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.5 | 3.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.5 | 1.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 2.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.5 | 3.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.5 | 3.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.5 | 10.9 | GO:0008306 | associative learning(GO:0008306) |
| 0.5 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 8.3 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.5 | 10.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.5 | 3.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.5 | 1.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.5 | 2.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.5 | 2.3 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.5 | 12.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.4 | 1.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 4.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 5.5 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.4 | 15.1 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.4 | 89.5 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.4 | 4.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.4 | 15.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.4 | 18.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.4 | 8.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 22.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.4 | 5.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.4 | 14.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 2.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 2.9 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.4 | 4.6 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 1.0 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.3 | 1.3 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 8.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 21.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.3 | 5.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.3 | 4.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 1.2 | GO:0045821 | transformation of host cell by virus(GO:0019087) positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.3 | 6.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 13.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.3 | 7.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.3 | 5.2 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.3 | 9.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.3 | 3.2 | GO:0021533 | cell differentiation in hindbrain(GO:0021533) |
| 0.3 | 8.5 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.3 | 0.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.3 | 2.5 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.3 | 0.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 2.4 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.3 | 3.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 11.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.3 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 104.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 2.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 11.3 | GO:0097581 | lamellipodium organization(GO:0097581) |
| 0.2 | 3.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 2.9 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 1.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 3.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 2.9 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.2 | 4.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.3 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 2.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 6.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 6.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 13.0 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 1.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 5.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.3 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.3 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.1 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.8 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 0.3 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 2.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 2.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.6 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.9 | 74.7 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 21.2 | 212.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 16.9 | 50.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 16.0 | 48.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 12.5 | 149.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 9.1 | 81.6 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 8.5 | 136.6 | GO:0031045 | dense core granule(GO:0031045) |
| 7.7 | 23.2 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 7.2 | 21.5 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 6.8 | 40.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 5.9 | 76.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 5.9 | 201.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 5.8 | 23.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 5.2 | 15.5 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 5.1 | 177.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 4.6 | 23.2 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 4.6 | 9.3 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 4.5 | 13.6 | GO:0060187 | cell pole(GO:0060187) |
| 4.0 | 92.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 3.9 | 121.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 3.7 | 29.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 3.7 | 44.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 3.6 | 21.5 | GO:0030478 | actin cap(GO:0030478) |
| 3.5 | 88.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 3.5 | 14.1 | GO:0034657 | GID complex(GO:0034657) |
| 3.5 | 10.5 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 3.5 | 27.7 | GO:0043203 | axon hillock(GO:0043203) |
| 3.3 | 40.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 3.2 | 105.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 3.1 | 21.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 3.0 | 12.1 | GO:0097196 | Shu complex(GO:0097196) |
| 3.0 | 27.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 2.8 | 16.9 | GO:0008091 | spectrin(GO:0008091) |
| 2.8 | 11.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 2.8 | 69.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 2.6 | 21.1 | GO:0001740 | Barr body(GO:0001740) |
| 2.5 | 25.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 2.4 | 12.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.4 | 99.9 | GO:0016528 | sarcoplasm(GO:0016528) |
| 2.3 | 27.7 | GO:0071437 | invadopodium(GO:0071437) |
| 2.2 | 13.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 2.2 | 50.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 2.1 | 2.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 2.1 | 33.1 | GO:0045180 | basal cortex(GO:0045180) |
| 2.0 | 6.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.0 | 10.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.9 | 78.8 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 1.9 | 9.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.8 | 14.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.8 | 1.8 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.8 | 14.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.8 | 47.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.7 | 10.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.7 | 54.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.6 | 52.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.6 | 9.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.6 | 6.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 1.6 | 9.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.5 | 75.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 1.5 | 250.1 | GO:0043204 | perikaryon(GO:0043204) |
| 1.4 | 225.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 1.4 | 17.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.4 | 12.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.4 | 30.7 | GO:0000145 | exocyst(GO:0000145) |
| 1.3 | 6.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.3 | 5.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.3 | 56.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 1.3 | 9.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.3 | 9.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.3 | 29.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 1.3 | 6.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.3 | 15.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.3 | 12.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 1.3 | 186.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 1.2 | 2.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 1.2 | 4.7 | GO:0032044 | DSIF complex(GO:0032044) |
| 1.2 | 43.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.2 | 10.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 1.1 | 5.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.1 | 10.0 | GO:0032059 | bleb(GO:0032059) |
| 1.1 | 41.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 1.1 | 3.3 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.1 | 5.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 73.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 1.1 | 126.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 1.0 | 29.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.0 | 21.2 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 1.0 | 12.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 63.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.9 | 21.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.9 | 30.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.9 | 18.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.8 | 11.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.8 | 109.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.8 | 55.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.8 | 4.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.8 | 5.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.7 | 83.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.7 | 7.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.7 | 17.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.7 | 4.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.7 | 2.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.7 | 16.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.6 | 4.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.6 | 45.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.6 | 1.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.6 | 3.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 10.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.6 | 23.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.6 | 22.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.6 | 31.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.6 | 4.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.5 | 3.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.5 | 4.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.5 | 18.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.5 | 11.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 5.9 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 28.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.4 | 1.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 3.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 5.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 33.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.3 | 39.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.3 | 2.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 3.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.3 | 36.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 5.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.3 | 3.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 6.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 4.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 3.8 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 10.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 7.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 106.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 2.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 3.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 28.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 10.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 2.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 8.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 639.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.6 | 154.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 31.0 | 92.9 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 18.2 | 54.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 14.8 | 73.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 14.4 | 43.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 14.0 | 56.0 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 13.8 | 55.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 13.6 | 81.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 12.7 | 50.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 12.2 | 36.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 11.6 | 46.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 11.4 | 79.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 10.6 | 31.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 10.1 | 60.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 9.5 | 75.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 9.4 | 65.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 9.2 | 46.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 9.1 | 27.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 8.8 | 26.5 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 8.5 | 50.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 8.3 | 74.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 8.0 | 39.9 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 7.9 | 47.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 7.8 | 23.5 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 7.8 | 23.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 7.5 | 37.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 7.5 | 29.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 7.4 | 22.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 7.4 | 7.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 7.2 | 21.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 7.1 | 28.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 7.0 | 21.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 6.9 | 20.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 6.8 | 20.5 | GO:2001070 | starch binding(GO:2001070) |
| 6.8 | 27.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 6.4 | 63.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 6.3 | 37.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 5.9 | 41.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 5.8 | 75.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 5.8 | 40.7 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 5.8 | 23.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 5.6 | 5.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 5.5 | 27.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 5.4 | 32.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 5.3 | 21.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 5.1 | 87.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 5.0 | 24.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 4.9 | 24.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 4.9 | 29.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 4.9 | 112.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 4.8 | 57.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 4.6 | 32.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 4.5 | 40.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 4.4 | 62.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 4.2 | 29.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 4.2 | 21.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 4.2 | 29.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 4.1 | 28.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 3.9 | 11.6 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 3.8 | 22.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 3.8 | 34.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 3.7 | 25.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 3.7 | 36.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 3.6 | 25.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 3.6 | 121.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 3.6 | 17.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 3.5 | 28.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 3.4 | 44.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 3.4 | 13.6 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 3.3 | 30.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 3.3 | 59.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 3.3 | 9.8 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 3.1 | 9.3 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 3.1 | 27.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 3.1 | 21.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 3.0 | 21.3 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 3.0 | 9.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 3.0 | 8.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 2.8 | 11.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 2.8 | 19.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 2.7 | 5.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 2.7 | 24.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.7 | 10.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 2.6 | 13.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 2.6 | 164.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 2.6 | 13.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 2.5 | 10.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 2.5 | 10.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 2.5 | 15.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.5 | 17.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 2.5 | 5.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 2.4 | 9.8 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 2.4 | 19.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 2.4 | 63.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 2.3 | 13.8 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 2.3 | 39.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 2.3 | 22.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 2.2 | 11.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 2.2 | 13.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 2.2 | 50.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 2.2 | 8.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.1 | 38.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 2.1 | 12.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 2.0 | 14.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 2.0 | 4.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 2.0 | 10.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 2.0 | 60.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 2.0 | 5.9 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) threonine-tRNA ligase activity(GO:0004829) |
| 2.0 | 23.6 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 2.0 | 21.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 2.0 | 5.9 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 1.9 | 13.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.8 | 9.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 1.8 | 11.0 | GO:0036122 | BMP binding(GO:0036122) |
| 1.8 | 14.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.8 | 59.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 1.7 | 17.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.7 | 5.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.7 | 10.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.7 | 20.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.7 | 6.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.7 | 10.0 | GO:0043495 | protein anchor(GO:0043495) |
| 1.6 | 100.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 1.6 | 14.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.6 | 7.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.5 | 6.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 1.5 | 22.9 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 1.4 | 40.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.4 | 17.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.4 | 9.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 1.4 | 7.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.4 | 9.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.4 | 23.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.3 | 23.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.3 | 7.8 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 1.3 | 97.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 1.3 | 28.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 1.3 | 30.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 1.3 | 3.8 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 1.3 | 27.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 1.3 | 6.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.2 | 11.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 1.2 | 46.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 1.1 | 25.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 1.1 | 12.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.1 | 13.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 1.0 | 8.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.0 | 10.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.0 | 47.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 1.0 | 8.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.0 | 8.8 | GO:0015266 | protein channel activity(GO:0015266) |
| 1.0 | 2.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.9 | 10.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.9 | 7.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.9 | 5.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.9 | 11.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.9 | 8.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.9 | 12.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.9 | 5.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.9 | 21.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.9 | 4.5 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.9 | 9.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.8 | 3.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.8 | 49.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.8 | 5.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.8 | 6.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.8 | 11.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.8 | 2.3 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 0.7 | 5.1 | GO:1904047 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.7 | 6.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.7 | 41.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.7 | 2.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.7 | 7.1 | GO:0016909 | JUN kinase activity(GO:0004705) cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) SAP kinase activity(GO:0016909) |
| 0.7 | 67.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.7 | 15.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.7 | 21.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 23.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.7 | 10.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.7 | 17.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.7 | 24.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.7 | 6.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 11.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 1.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.6 | 9.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.6 | 12.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.6 | 8.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 15.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 2.5 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.6 | 1.8 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.6 | 10.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.6 | 6.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 25.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.6 | 8.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.6 | 2.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 10.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 5.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.5 | 6.8 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.5 | 23.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.5 | 19.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.5 | 1.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.5 | 6.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 51.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 12.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.5 | 5.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 52.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 9.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 8.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.4 | 18.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.4 | 4.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 40.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.4 | 11.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.4 | 111.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.4 | 5.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.4 | 89.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.4 | 31.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.4 | 1.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 3.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 8.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 4.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 1.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 2.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 10.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 42.4 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.3 | 1.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 13.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 15.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 6.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 4.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.8 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.3 | 2.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.3 | 3.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 4.5 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 3.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.3 | 2.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 4.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 7.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 2.8 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.2 | 27.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 12.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 16.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 3.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 3.5 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.2 | 6.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 3.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 3.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 2.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 6.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 4.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 2.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 3.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 55.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 6.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 3.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 24.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 5.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 5.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 24.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 4.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 4.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 119.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 3.5 | 13.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 3.1 | 122.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 3.0 | 62.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 2.2 | 26.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 2.0 | 27.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.9 | 70.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 1.8 | 7.1 | PID FOXO PATHWAY | FoxO family signaling |
| 1.8 | 47.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 1.4 | 16.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 1.2 | 57.6 | PID BMP PATHWAY | BMP receptor signaling |
| 1.2 | 42.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 1.2 | 72.9 | PID FGF PATHWAY | FGF signaling pathway |
| 1.1 | 14.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 1.0 | 29.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 1.0 | 9.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.0 | 36.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 1.0 | 31.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 1.0 | 23.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 1.0 | 13.7 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 1.0 | 13.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.9 | 7.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.9 | 50.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.8 | 5.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.8 | 19.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.8 | 188.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.8 | 115.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.6 | 10.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.6 | 25.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.6 | 17.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.5 | 10.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 114.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.5 | 5.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 15.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 5.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.5 | 7.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 8.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.4 | 9.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 5.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 9.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.4 | 7.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 2.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 5.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.3 | 7.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 2.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 3.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 4.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 1.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 110.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 8.0 | 120.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 6.8 | 115.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 6.8 | 121.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 5.1 | 126.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 4.9 | 88.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 4.2 | 62.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 3.8 | 98.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 3.5 | 52.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 3.2 | 9.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 3.0 | 47.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 2.9 | 35.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 2.6 | 12.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 2.2 | 82.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 2.2 | 31.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 2.1 | 20.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 2.1 | 24.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 2.0 | 21.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 1.9 | 40.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.9 | 53.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.9 | 74.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 1.8 | 54.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.8 | 125.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 1.7 | 30.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 1.6 | 30.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 1.6 | 44.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 1.6 | 20.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.5 | 22.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.5 | 13.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 1.5 | 22.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.5 | 4.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.4 | 34.4 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 1.4 | 22.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 1.4 | 15.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.3 | 34.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 1.2 | 117.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 1.2 | 8.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 1.1 | 19.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 1.1 | 32.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.0 | 39.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 1.0 | 18.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.0 | 42.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 1.0 | 7.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 1.0 | 21.4 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 1.0 | 32.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 1.0 | 17.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.9 | 5.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.9 | 27.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.9 | 20.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.9 | 30.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.8 | 9.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 7.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.8 | 70.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.7 | 11.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.7 | 120.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.7 | 55.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.7 | 7.8 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.7 | 9.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.7 | 8.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 8.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.6 | 6.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.6 | 22.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.6 | 5.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.6 | 9.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 5.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.5 | 12.4 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.5 | 11.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 9.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.5 | 2.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.5 | 9.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.4 | 6.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 6.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 3.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 3.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 22.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 8.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 8.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 14.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 0.6 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |