Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
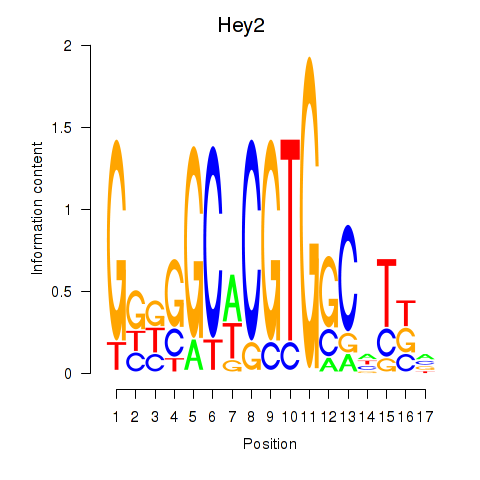
Results for Hey2
Z-value: 0.64
Transcription factors associated with Hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hey2
|
ENSRNOG00000013364 | hes-related family bHLH transcription factor with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | rn6_v1_chr1_+_29191192_29191192 | 0.53 | 2.2e-24 | Click! |
Activity profile of Hey2 motif
Sorted Z-values of Hey2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_26906716 | 34.00 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr3_-_9262628 | 19.87 |
ENSRNOT00000013286
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr8_+_72405748 | 15.54 |
ENSRNOT00000023952
|
Car12
|
carbonic anhydrase 12 |
| chr11_+_81972219 | 15.22 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr19_+_37476095 | 13.78 |
ENSRNOT00000092794
ENSRNOT00000023130 |
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr1_+_101682172 | 12.54 |
ENSRNOT00000028540
|
Car11
|
carbonic anhydrase 11 |
| chr3_-_146470293 | 11.74 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_-_108210826 | 11.45 |
ENSRNOT00000064705
|
Dock9
|
dedicator of cytokinesis 9 |
| chr16_+_48120864 | 11.02 |
ENSRNOT00000068562
|
Stox2
|
storkhead box 2 |
| chr13_-_111765944 | 10.54 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr9_+_41348449 | 10.48 |
ENSRNOT00000019419
|
Plekhb2
|
pleckstrin homology domain containing B2 |
| chr2_-_46544457 | 10.26 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr14_+_89253373 | 9.99 |
ENSRNOT00000035922
|
LOC688553
|
hypothetical protein LOC688553 |
| chr2_+_115337439 | 9.57 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr14_-_92117923 | 9.11 |
ENSRNOT00000044726
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_+_39711201 | 9.01 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr2_+_207923775 | 8.97 |
ENSRNOT00000019997
ENSRNOT00000051835 |
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr6_-_28464118 | 8.77 |
ENSRNOT00000068214
|
Efr3b
|
EFR3 homolog B |
| chr1_-_47331412 | 8.67 |
ENSRNOT00000046746
|
Ezr
|
ezrin |
| chr12_+_39553903 | 8.64 |
ENSRNOT00000001738
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr2_-_197991198 | 8.49 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr12_+_39554171 | 8.41 |
ENSRNOT00000024347
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr4_-_120817377 | 8.01 |
ENSRNOT00000021826
|
Podxl2
|
podocalyxin-like 2 |
| chr14_-_10395047 | 7.55 |
ENSRNOT00000002936
|
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr16_-_21347968 | 7.24 |
ENSRNOT00000068554
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr9_-_45505767 | 6.88 |
ENSRNOT00000033964
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr10_+_47961056 | 6.75 |
ENSRNOT00000027312
|
Fam83g
|
family with sequence similarity 83, member G |
| chr2_-_197991574 | 6.62 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr7_+_97559841 | 6.55 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr8_+_65686648 | 6.34 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr10_+_92018562 | 5.99 |
ENSRNOT00000006483
|
Arf2
|
ADP-ribosylation factor 2 |
| chr9_+_98073038 | 5.69 |
ENSRNOT00000026795
|
Mlph
|
melanophilin |
| chr5_-_25584278 | 5.49 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_-_82452281 | 5.44 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr12_-_30566032 | 5.31 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr15_+_18399515 | 5.26 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr8_+_5893249 | 5.15 |
ENSRNOT00000014041
|
Mmp7
|
matrix metallopeptidase 7 |
| chr8_+_40001030 | 5.08 |
ENSRNOT00000083778
|
Esam
|
endothelial cell adhesion molecule |
| chr19_-_44158621 | 5.05 |
ENSRNOT00000031243
ENSRNOT00000088297 |
Tmem231
|
transmembrane protein 231 |
| chr13_-_70174565 | 5.02 |
ENSRNOT00000067135
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr2_+_142262236 | 4.98 |
ENSRNOT00000019057
|
Lhfp
|
lipoma HMGIC fusion partner |
| chr3_-_82236664 | 4.94 |
ENSRNOT00000011650
|
Tspan18
|
tetraspanin 18 |
| chr13_-_90832138 | 4.92 |
ENSRNOT00000010930
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr14_+_77712240 | 4.78 |
ENSRNOT00000009101
|
Msx1
|
msh homeobox 1 |
| chr16_+_48121430 | 4.71 |
ENSRNOT00000090839
|
Stox2
|
storkhead box 2 |
| chr14_+_7169519 | 4.69 |
ENSRNOT00000003026
|
Klhl8
|
kelch-like family member 8 |
| chr17_-_15511972 | 4.58 |
ENSRNOT00000074041
|
Aspnl1
|
asporin-like 1 |
| chr14_-_80169431 | 4.51 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr7_+_94795214 | 4.50 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr17_-_13812615 | 4.41 |
ENSRNOT00000019473
ENSRNOT00000085581 |
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr7_-_126913585 | 4.35 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr11_+_32211115 | 4.33 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr10_-_82852660 | 4.33 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chr16_-_21348391 | 4.29 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr10_+_3338808 | 3.97 |
ENSRNOT00000004384
ENSRNOT00000093444 |
Mpv17l
|
MPV17 mitochondrial inner membrane protein like |
| chr5_-_161035368 | 3.96 |
ENSRNOT00000091640
|
AABR07050321.2
|
|
| chr4_-_66002444 | 3.51 |
ENSRNOT00000018645
|
Zc3hav1l
|
zinc finger CCCH-type containing, antiviral 1 like |
| chr6_+_110624856 | 3.46 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr7_+_27309966 | 3.30 |
ENSRNOT00000031871
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr9_-_113900190 | 3.18 |
ENSRNOT00000016965
|
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr8_-_47393503 | 3.12 |
ENSRNOT00000059868
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr17_+_33408722 | 2.99 |
ENSRNOT00000023691
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr1_+_113034227 | 2.90 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr9_+_80118029 | 2.74 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr16_+_9563218 | 2.73 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr18_-_56534415 | 2.71 |
ENSRNOT00000024374
|
Slc26a2
|
solute carrier family 26 member 2 |
| chr7_+_122979021 | 2.66 |
ENSRNOT00000085707
|
Zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr13_-_76049363 | 2.45 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr3_+_58084606 | 2.42 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr17_-_6244612 | 2.41 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr7_-_123088279 | 2.40 |
ENSRNOT00000071998
|
Tob2
|
transducer of ERBB2, 2 |
| chr3_-_51297852 | 2.33 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr1_-_87937516 | 2.29 |
ENSRNOT00000087522
|
Eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr5_-_160070916 | 2.24 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr9_-_15393131 | 2.22 |
ENSRNOT00000018523
ENSRNOT00000088314 |
Med20
|
mediator complex subunit 20 |
| chr13_-_90832469 | 2.13 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr10_-_61744976 | 2.10 |
ENSRNOT00000079926
ENSRNOT00000092314 ENSRNOT00000034298 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr5_+_90419020 | 2.10 |
ENSRNOT00000058893
|
Rasef
|
RAS and EF hand domain containing |
| chr6_-_109162267 | 2.03 |
ENSRNOT00000077518
|
Nek9
|
NIMA-related kinase 9 |
| chr7_+_77678968 | 1.98 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr3_-_168033457 | 1.98 |
ENSRNOT00000055111
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_+_61685241 | 1.97 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr3_+_119698652 | 1.96 |
ENSRNOT00000088834
ENSRNOT00000066114 |
Stard7
|
StAR-related lipid transfer domain containing 7 |
| chr15_+_33108671 | 1.92 |
ENSRNOT00000015468
|
Lrp10
|
LDL receptor related protein 10 |
| chr1_+_221673590 | 1.91 |
ENSRNOT00000038016
|
Cdc42bpg
|
CDC42 binding protein kinase gamma |
| chr5_+_156215417 | 1.88 |
ENSRNOT00000067616
ENSRNOT00000077742 |
Ece1
|
endothelin converting enzyme 1 |
| chr8_+_56179637 | 1.85 |
ENSRNOT00000035989
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr19_+_41710102 | 1.84 |
ENSRNOT00000021866
ENSRNOT00000079730 |
Marveld3
|
MARVEL domain containing 3 |
| chr3_+_164822111 | 1.66 |
ENSRNOT00000014568
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr12_+_24761210 | 1.60 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr14_-_11198194 | 1.57 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr5_+_135735825 | 1.52 |
ENSRNOT00000068267
|
Zswim5
|
zinc finger, SWIM-type containing 5 |
| chr9_+_20213776 | 1.42 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr10_-_81653717 | 1.37 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr3_-_156905892 | 1.32 |
ENSRNOT00000022424
|
Emilin3
|
elastin microfibril interfacer 3 |
| chr16_+_71787966 | 1.29 |
ENSRNOT00000080084
|
Htra4
|
HtrA serine peptidase 4 |
| chr1_+_86914137 | 1.19 |
ENSRNOT00000027006
|
Fbxo17
|
F-box protein 17 |
| chr2_-_143104412 | 1.19 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr15_-_33589522 | 1.08 |
ENSRNOT00000022514
|
Efs
|
embryonal Fyn-associated substrate |
| chr7_-_12779862 | 0.91 |
ENSRNOT00000080292
|
Tmem259
|
transmembrane protein 259 |
| chr14_+_85351358 | 0.88 |
ENSRNOT00000031914
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr16_+_73564243 | 0.88 |
ENSRNOT00000030960
|
Golga7
|
golgin A7 |
| chr7_-_11330278 | 0.86 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr15_+_58554374 | 0.84 |
ENSRNOT00000058206
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr3_+_175408629 | 0.80 |
ENSRNOT00000081344
|
Lsm14b
|
LSM family member 14B |
| chr7_+_80351774 | 0.73 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chr3_+_134413170 | 0.53 |
ENSRNOT00000074338
|
AABR07054000.1
|
|
| chr5_-_173339890 | 0.50 |
ENSRNOT00000067704
ENSRNOT00000078453 |
Pusl1
|
pseudouridylate synthase-like 1 |
| chr4_-_98593664 | 0.50 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr9_-_37231291 | 0.50 |
ENSRNOT00000016237
|
Ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr9_+_66495832 | 0.39 |
ENSRNOT00000022676
ENSRNOT00000091283 |
Nop58
|
NOP58 ribonucleoprotein |
| chr1_-_221087449 | 0.35 |
ENSRNOT00000016987
|
Fam89b
|
family with sequence similarity 89, member B |
| chr6_+_73345392 | 0.33 |
ENSRNOT00000088378
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr1_-_112947399 | 0.24 |
ENSRNOT00000093306
ENSRNOT00000093259 |
Gabra5
|
gamma-aminobutyric acid type A receptor alpha 5 subunit |
| chr1_-_200163106 | 0.19 |
ENSRNOT00000027644
|
Mcmbp
|
minichromosome maintenance complex binding protein |
| chr10_+_62674561 | 0.17 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chr6_-_92527711 | 0.13 |
ENSRNOT00000007529
|
Nin
|
ninein |
| chr7_-_97944491 | 0.11 |
ENSRNOT00000008454
|
Zhx1
|
zinc fingers and homeoboxes 1 |
| chr19_+_25123724 | 0.08 |
ENSRNOT00000007407
|
LOC686013
|
hypothetical protein LOC686013 |
| chr4_+_6559545 | 0.03 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr9_+_94880053 | 0.01 |
ENSRNOT00000024445
|
Atg16l1
|
autophagy related 16-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 34.0 | GO:0030070 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 5.7 | 17.0 | GO:1903233 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 3.9 | 11.7 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 2.2 | 8.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.7 | 5.2 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 1.6 | 9.6 | GO:0045901 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 1.6 | 4.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 1.6 | 15.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.5 | 10.3 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.5 | 4.4 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.1 | 4.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.0 | 3.0 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 1.0 | 4.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.9 | 3.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.9 | 13.8 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.8 | 7.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.8 | 2.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.7 | 9.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.7 | 2.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 9.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.6 | 1.9 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.6 | 15.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.5 | 15.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.4 | 5.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.4 | 11.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.4 | 4.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.4 | 5.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 4.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 6.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.3 | 2.4 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.3 | 15.7 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.3 | 1.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 8.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 1.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 5.7 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.2 | 2.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 10.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.4 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.2 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 2.7 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 3.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.9 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 4.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 12.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 5.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 1.4 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 4.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.9 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 2.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 6.7 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 2.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 4.2 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 2.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 13.4 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 1.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.2 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 1.6 | GO:0048839 | inner ear development(GO:0048839) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 2.2 | 8.7 | GO:0036398 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 2.1 | 34.0 | GO:0031045 | dense core granule(GO:0031045) |
| 1.4 | 4.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.3 | 6.3 | GO:0043293 | apoptosome(GO:0043293) |
| 1.1 | 5.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.6 | 5.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 9.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 2.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 1.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 5.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.3 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 9.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 19.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 4.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 14.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 7.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 5.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 7.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.5 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 2.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0045177 | apical part of cell(GO:0045177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.9 | 11.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.8 | 34.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.8 | 5.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 1.8 | 9.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.4 | 25.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.3 | 7.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.2 | 15.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 1.1 | 15.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 1.1 | 4.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 5.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.8 | 10.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.6 | 5.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.6 | 3.0 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.5 | 2.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.5 | 1.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.4 | 4.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 9.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 2.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 9.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 15.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 7.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.4 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.2 | 10.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 12.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 11.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 8.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 7.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 6.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 4.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0001948 | glycoprotein binding(GO:0001948) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 33.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.4 | 11.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 8.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 8.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 9.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 10.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 10.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 4.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 7.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.3 | 33.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.9 | 17.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.7 | 9.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 9.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 15.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 9.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 5.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 8.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 5.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 9.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 11.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 7.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 3.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 4.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |