Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
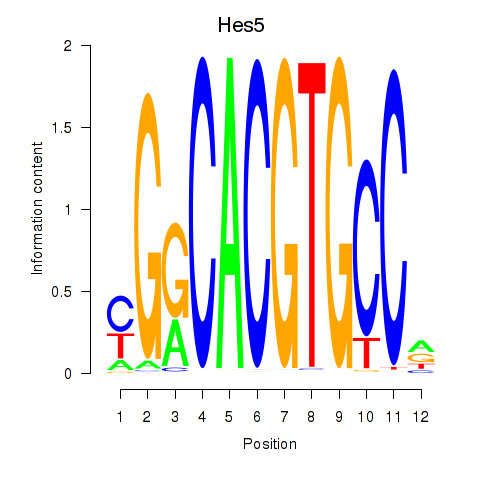
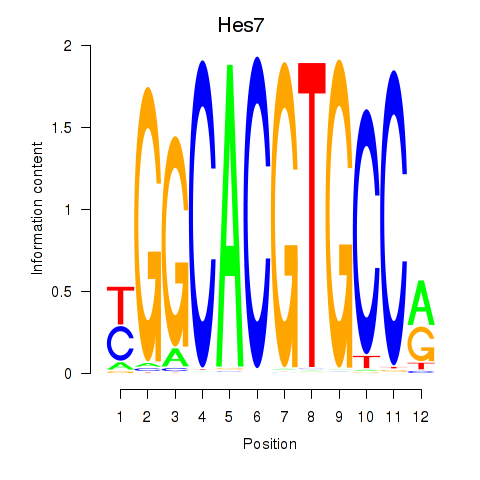
Results for Hes5_Hes7
Z-value: 0.70
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSRNOG00000013850 | hes family bHLH transcription factor 5 |
|
Hes7
|
ENSRNOG00000007391 | hes family bHLH transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes7 | rn6_v1_chr10_+_55707164_55707164 | 0.46 | 2.1e-18 | Click! |
| Hes5 | rn6_v1_chr5_+_172364421_172364421 | -0.26 | 1.7e-06 | Click! |
Activity profile of Hes5_Hes7 motif
Sorted Z-values of Hes5_Hes7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92117923 | 106.59 |
ENSRNOT00000044726
|
Grb10
|
growth factor receptor bound protein 10 |
| chr2_+_92549479 | 27.14 |
ENSRNOT00000082912
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr8_+_64489942 | 23.32 |
ENSRNOT00000083666
|
Pkm
|
pyruvate kinase, muscle |
| chr1_+_190072420 | 22.48 |
ENSRNOT00000037026
|
Abca15
|
ATP-binding cassette, subfamily A (ABC1), member 15 |
| chr8_+_128972311 | 22.31 |
ENSRNOT00000025460
|
Myrip
|
myosin VIIA and Rab interacting protein |
| chr2_+_197682000 | 21.74 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr1_-_220787238 | 17.38 |
ENSRNOT00000083656
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr13_-_111765944 | 17.01 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr2_+_147006830 | 16.45 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr7_+_38945836 | 15.81 |
ENSRNOT00000006455
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr2_-_196415530 | 15.21 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr4_-_157331905 | 15.16 |
ENSRNOT00000020647
|
Tpi1
|
triosephosphate isomerase 1 |
| chr10_-_82852660 | 14.66 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chr9_+_20213776 | 13.98 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr4_-_58216711 | 13.23 |
ENSRNOT00000015134
|
Tsga13
|
testis specific, 13 |
| chr16_+_71889235 | 13.09 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr6_-_26486695 | 12.77 |
ENSRNOT00000073236
|
Krtcap3
|
keratinocyte associated protein 3 |
| chr19_-_25134055 | 12.74 |
ENSRNOT00000007519
|
Misp3
|
MISP family member 3 |
| chr20_-_5754773 | 11.26 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr16_-_73827488 | 11.05 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr4_-_82194927 | 11.02 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr3_+_152143811 | 9.47 |
ENSRNOT00000026578
|
LOC100911109
|
sperm-associated antigen 4 protein-like |
| chr7_+_77966722 | 9.13 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr5_-_27180819 | 8.97 |
ENSRNOT00000045483
|
AABR07047293.1
|
|
| chr10_-_15960673 | 8.41 |
ENSRNOT00000036447
|
RGD1311343
|
similar to RIKEN cDNA 4930524B15 |
| chr6_-_59950586 | 8.38 |
ENSRNOT00000005800
|
Arl4a
|
ADP-ribosylation factor like GTPase 4A |
| chr10_-_36716601 | 8.30 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr14_-_114868328 | 8.06 |
ENSRNOT00000034584
|
RGD1562229
|
similar to hypothetical protein FLJ40298 |
| chr2_+_85377318 | 8.03 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr10_-_74119009 | 7.83 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr1_-_164590562 | 7.66 |
ENSRNOT00000024157
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr3_-_92290919 | 6.89 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr8_-_36760742 | 6.79 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr5_-_160158386 | 6.76 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr5_-_156141537 | 6.71 |
ENSRNOT00000019004
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr4_-_155561324 | 6.35 |
ENSRNOT00000085447
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr5_-_136112344 | 5.74 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr17_-_13393243 | 5.73 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr10_-_46145548 | 5.45 |
ENSRNOT00000033483
|
Pld6
|
phospholipase D family, member 6 |
| chr3_+_56030915 | 5.24 |
ENSRNOT00000010489
|
Phospho2
|
phosphatase, orphan 2 |
| chr10_-_12916784 | 5.10 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr3_+_148082790 | 5.04 |
ENSRNOT00000009930
ENSRNOT00000080767 |
Defb36
|
defensin beta 36 |
| chr7_-_12905339 | 4.80 |
ENSRNOT00000088330
|
Cdc34
|
cell division cycle 34 |
| chr7_+_77899322 | 4.72 |
ENSRNOT00000006195
|
Fzd6
|
frizzled class receptor 6 |
| chr4_-_157486844 | 4.54 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr20_-_13458110 | 4.47 |
ENSRNOT00000072183
|
Slc5a4
|
solute carrier family 5 member 4 |
| chr1_+_265883355 | 4.46 |
ENSRNOT00000025938
|
Elovl3
|
ELOVL fatty acid elongase 3 |
| chr3_-_56030744 | 4.42 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr3_+_138770574 | 4.18 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr9_+_118842787 | 4.13 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr5_-_156105867 | 4.00 |
ENSRNOT00000078851
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr2_+_211176556 | 3.93 |
ENSRNOT00000055880
|
Psrc1
|
proline and serine rich coiled-coil 1 |
| chr1_+_185673177 | 3.83 |
ENSRNOT00000048020
|
Sox6
|
SRY box 6 |
| chr11_+_24266345 | 3.66 |
ENSRNOT00000046973
|
Jam2
|
junctional adhesion molecule 2 |
| chr17_-_56909992 | 3.56 |
ENSRNOT00000021801
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chrX_-_15520712 | 3.52 |
ENSRNOT00000011994
|
Kcnd1
|
potassium voltage-gated channel subfamily D member 1 |
| chr5_-_117612123 | 3.37 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr2_+_189400696 | 3.31 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr9_-_82419288 | 3.24 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr3_-_92242318 | 3.22 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr16_-_21348391 | 3.16 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr16_-_755990 | 3.09 |
ENSRNOT00000013501
|
Polr3a
|
RNA polymerase III subunit A |
| chr14_+_7171613 | 3.00 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr11_+_47090245 | 2.95 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr4_-_157679962 | 2.95 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr19_-_25914689 | 2.95 |
ENSRNOT00000091994
ENSRNOT00000039354 |
Nf1x
|
nuclear factor 1 X |
| chr16_-_21347968 | 2.72 |
ENSRNOT00000068554
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr16_+_19068783 | 2.67 |
ENSRNOT00000017424
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr7_+_63094911 | 2.57 |
ENSRNOT00000006426
|
Wif1
|
Wnt inhibitory factor 1 |
| chr10_-_84789832 | 2.47 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_-_189400323 | 2.43 |
ENSRNOT00000024364
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr5_+_60809762 | 2.32 |
ENSRNOT00000016839
|
Frmpd1
|
FERM and PDZ domain containing 1 |
| chr1_+_220307394 | 2.25 |
ENSRNOT00000071891
|
LOC108348052
|
equilibrative nucleoside transporter 2 |
| chr20_-_43108198 | 2.24 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr5_+_152681101 | 2.23 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr10_+_45659143 | 2.17 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr16_+_83206004 | 2.11 |
ENSRNOT00000018870
|
Ankrd10
|
ankyrin repeat domain 10 |
| chr1_+_82089922 | 1.97 |
ENSRNOT00000071251
|
Zfp526
|
zinc finger protein 526 |
| chr1_-_169598835 | 1.90 |
ENSRNOT00000072996
|
Olr156
|
olfactory receptor 156 |
| chr10_+_35133252 | 1.89 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr5_+_128215711 | 1.86 |
ENSRNOT00000011670
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr7_-_12779862 | 1.84 |
ENSRNOT00000080292
|
Tmem259
|
transmembrane protein 259 |
| chrY_+_151937 | 1.80 |
ENSRNOT00000077988
ENSRNOT00000083382 |
Tspy1
|
testis specific protein, Y-linked 1 |
| chr18_+_63599425 | 1.80 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr7_+_126096793 | 1.78 |
ENSRNOT00000047234
ENSRNOT00000088042 |
Fbln1
|
fibulin 1 |
| chr20_+_7718282 | 1.73 |
ENSRNOT00000000593
|
Scube3
|
signal peptide, CUB domain and EGF like domain containing 3 |
| chr12_+_52359310 | 1.68 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr7_+_77678968 | 1.48 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr1_+_78546012 | 1.45 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chr1_+_102017263 | 1.44 |
ENSRNOT00000028680
|
Nomo1
|
nodal modulator 1 |
| chr2_+_95320283 | 1.37 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr5_+_150459713 | 1.37 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr2_-_93641497 | 1.33 |
ENSRNOT00000013720
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr1_+_148862493 | 1.09 |
ENSRNOT00000073451
|
LOC100912097
|
olfactory receptor 10A5-like |
| chr10_+_91710495 | 1.02 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr16_-_9709347 | 0.89 |
ENSRNOT00000083933
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr3_+_125503638 | 0.78 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr2_+_40000313 | 0.77 |
ENSRNOT00000014270
|
Depdc1b
|
DEP domain containing 1B |
| chr1_-_282251257 | 0.75 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr9_+_80118029 | 0.65 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr18_+_72005581 | 0.63 |
ENSRNOT00000072519
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr16_-_75855745 | 0.60 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr10_+_36530719 | 0.59 |
ENSRNOT00000046476
|
Olr1409
|
olfactory receptor 1409 |
| chr20_-_48758905 | 0.58 |
ENSRNOT00000000710
|
Ddo
|
D-aspartate oxidase |
| chr1_+_171095325 | 0.56 |
ENSRNOT00000026503
|
Olr227
|
olfactory receptor 227 |
| chrX_+_124228270 | 0.54 |
ENSRNOT00000035449
|
Rhox11
|
reproductive homeobox 11 |
| chr19_-_57422093 | 0.45 |
ENSRNOT00000025512
|
RGD1559896
|
similar to RIKEN cDNA 2310022B05 |
| chr3_+_73064092 | 0.36 |
ENSRNOT00000072178
|
LOC684539
|
similar to olfactory receptor Olr490 |
| chr20_+_4576057 | 0.33 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr3_+_138508899 | 0.32 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr8_-_43249887 | 0.30 |
ENSRNOT00000060094
|
Olr1305
|
olfactory receptor 1305 |
| chr2_+_150211898 | 0.28 |
ENSRNOT00000018767
|
Sucnr1
|
succinate receptor 1 |
| chr15_-_28446785 | 0.28 |
ENSRNOT00000015439
|
Olr1638
|
olfactory receptor 1638 |
| chr18_+_30398113 | 0.22 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr3_+_47439076 | 0.19 |
ENSRNOT00000011794
|
Tank
|
TRAF family member-associated NFKB activator |
| chr5_-_137617258 | 0.19 |
ENSRNOT00000071641
|
Olfr1330-ps1
|
olfactory receptor 1330, pseudogene 1 |
| chr20_+_11168298 | 0.17 |
ENSRNOT00000032240
|
Trappc10
|
trafficking protein particle complex 10 |
| chr10_+_10889488 | 0.17 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr1_+_224933920 | 0.15 |
ENSRNOT00000066823
|
Wdr74
|
WD repeat domain 74 |
| chr11_+_83883879 | 0.06 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr10_+_83476107 | 0.04 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr10_+_11338306 | 0.01 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr2_-_57600820 | 0.01 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes5_Hes7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.2 | 106.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 3.7 | 14.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 3.6 | 10.7 | GO:0071529 | cementum mineralization(GO:0071529) |
| 3.1 | 21.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 3.0 | 15.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 2.3 | 23.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 2.3 | 9.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 1.8 | 5.4 | GO:0007315 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 1.6 | 4.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 1.0 | 5.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 1.0 | 8.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 1.0 | 2.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.9 | 4.5 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 2.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.7 | 14.0 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.6 | 3.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 6.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.6 | 4.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.6 | 1.8 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.6 | 2.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 4.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.5 | 6.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.5 | 3.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.5 | 1.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.5 | 11.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.4 | 4.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 2.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.3 | 2.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 11.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 5.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 1.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.3 | 3.6 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.3 | 0.9 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.3 | 0.6 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 2.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 22.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 2.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 4.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 4.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 5.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 9.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 3.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 8.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 2.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 0.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 16.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 0.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.8 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 3.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 23.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 1.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 6.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 1.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 7.1 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 3.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.4 | 22.3 | GO:0031045 | dense core granule(GO:0031045) |
| 1.3 | 10.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 1.1 | 11.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 21.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.4 | 2.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 1.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 6.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 2.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 6.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 1.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 6.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 1.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 3.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 21.2 | GO:0061695 | transferase complex, transferring phosphorus-containing groups(GO:0061695) |
| 0.1 | 2.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 7.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 7.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 26.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 5.8 | 23.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 3.7 | 14.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 3.5 | 94.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 2.2 | 11.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 2.1 | 6.3 | GO:0033222 | xylose binding(GO:0033222) |
| 2.0 | 8.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.3 | 10.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.3 | 11.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 2.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.7 | 2.9 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.7 | 3.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.6 | 6.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 13.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.5 | 1.4 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 4.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 4.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 22.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.3 | 4.1 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 1.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 4.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 3.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 0.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 5.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 11.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 22.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 4.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 9.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 13.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 7.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 8.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 106.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.6 | 16.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 5.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 5.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 13.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 106.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 1.0 | 14.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 18.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 8.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 6.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 6.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 10.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 3.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 5.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |