Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
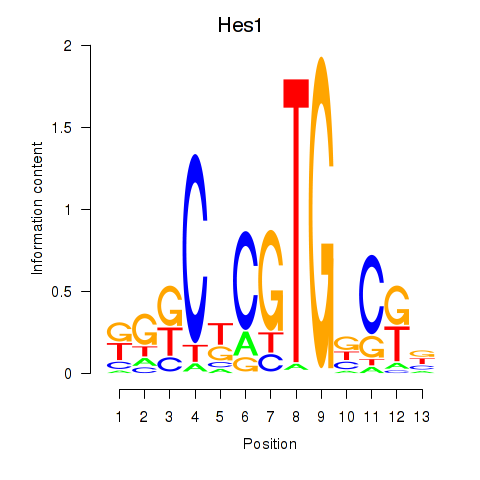
Results for Hes1
Z-value: 0.74
Transcription factors associated with Hes1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes1
|
ENSRNOG00000001720 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes1 | rn6_v1_chr11_-_74315248_74315248 | -0.50 | 4.9e-22 | Click! |
Activity profile of Hes1 motif
Sorted Z-values of Hes1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_153217782 | 13.58 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr14_-_86297623 | 12.86 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr13_-_98480419 | 12.64 |
ENSRNOT00000086306
|
Coq8a
|
coenzyme Q8A |
| chr2_+_207108552 | 11.89 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr8_+_48422036 | 10.71 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr3_+_150910398 | 10.57 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr8_+_48716939 | 10.50 |
ENSRNOT00000015810
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr7_-_119689938 | 10.45 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr7_+_126619196 | 9.15 |
ENSRNOT00000030082
|
Ppara
|
peroxisome proliferator activated receptor alpha |
| chr15_-_52029816 | 9.05 |
ENSRNOT00000013067
|
Slc39a14
|
solute carrier family 39 member 14 |
| chr12_+_39553903 | 8.59 |
ENSRNOT00000001738
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr2_-_260596777 | 8.59 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr5_+_156810991 | 8.05 |
ENSRNOT00000055904
|
AABR07050222.1
|
|
| chr15_-_27819376 | 8.03 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr10_-_109821807 | 8.00 |
ENSRNOT00000054949
|
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr1_-_143535583 | 7.99 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr10_-_29450644 | 7.67 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr7_-_70842405 | 7.33 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr9_+_10428853 | 7.31 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr19_+_11473541 | 7.29 |
ENSRNOT00000082066
|
Amfr
|
autocrine motility factor receptor |
| chr6_+_30638733 | 7.13 |
ENSRNOT00000082885
|
AABR07063424.1
|
rRNA promoter binding protein |
| chr4_+_10823281 | 7.12 |
ENSRNOT00000066739
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr10_+_40208223 | 7.01 |
ENSRNOT00000000772
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr2_-_22385676 | 6.89 |
ENSRNOT00000065224
|
Thbs4
|
thrombospondin 4 |
| chr2_-_22385855 | 6.81 |
ENSRNOT00000083531
|
Thbs4
|
thrombospondin 4 |
| chr20_-_10386663 | 6.77 |
ENSRNOT00000045275
ENSRNOT00000042432 |
Cbs
|
cystathionine beta synthase |
| chr2_+_127845034 | 6.75 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr10_-_62699723 | 6.60 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr5_-_152746588 | 6.47 |
ENSRNOT00000022908
|
Mtfr1l
|
mitochondrial fission regulator 1-like |
| chr11_-_38420032 | 6.37 |
ENSRNOT00000002209
ENSRNOT00000080681 |
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr9_-_82214440 | 6.12 |
ENSRNOT00000024419
|
Ihh
|
Indian hedgehog |
| chr9_+_47386626 | 6.10 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr1_-_265899958 | 6.02 |
ENSRNOT00000026013
|
Pitx3
|
paired-like homeodomain 3 |
| chr5_-_152589719 | 5.91 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr1_+_215460226 | 5.48 |
ENSRNOT00000027270
|
LOC685544
|
hypothetical protein LOC685544 |
| chr4_-_82229397 | 5.36 |
ENSRNOT00000089432
|
Hoxa13
|
homeo box A13 |
| chr1_-_263959318 | 5.26 |
ENSRNOT00000068007
|
Pkd2l1
|
polycystin 2 like 1, transient receptor potential cation channel |
| chr1_-_90091287 | 5.24 |
ENSRNOT00000032613
|
Gpi
|
glucose-6-phosphate isomerase |
| chr7_+_70452579 | 5.23 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chrX_+_156227830 | 5.10 |
ENSRNOT00000083949
|
LOC690348
|
similar to ESO3 protein |
| chr4_-_117296082 | 5.00 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr1_-_82452281 | 4.99 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr3_+_114176309 | 4.97 |
ENSRNOT00000023350
|
Sord
|
sorbitol dehydrogenase |
| chr1_+_166893734 | 4.96 |
ENSRNOT00000026702
|
Phox2a
|
paired-like homeobox 2a |
| chr8_-_111850393 | 4.94 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr3_+_154395187 | 4.89 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr3_+_147585947 | 4.88 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chr7_+_54980120 | 4.81 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr12_+_45026886 | 4.79 |
ENSRNOT00000001500
|
Pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr13_+_83972212 | 4.78 |
ENSRNOT00000004394
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr7_+_66595742 | 4.75 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr7_-_28040510 | 4.72 |
ENSRNOT00000005674
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr1_-_86948845 | 4.67 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr13_+_48745860 | 4.64 |
ENSRNOT00000010242
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr9_-_69953182 | 4.61 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr8_-_130491998 | 4.59 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr8_-_117353672 | 4.54 |
ENSRNOT00000027262
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr9_-_100624638 | 4.53 |
ENSRNOT00000051155
|
Hdlbp
|
high density lipoprotein binding protein |
| chr5_-_156811650 | 4.51 |
ENSRNOT00000068065
|
Fam43b
|
family with sequence similarity 43, member B |
| chr7_-_70452675 | 4.48 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr10_+_38814799 | 4.39 |
ENSRNOT00000074769
|
Shroom1
|
shroom family member 1 |
| chr7_-_18634079 | 4.31 |
ENSRNOT00000010031
|
Angptl4
|
angiopoietin-like 4 |
| chr5_-_148492232 | 4.09 |
ENSRNOT00000017577
|
Serinc2
|
serine incorporator 2 |
| chr10_-_53037816 | 3.99 |
ENSRNOT00000057509
|
Shisa6
|
shisa family member 6 |
| chr6_-_141008427 | 3.92 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chr3_+_13838304 | 3.87 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr10_+_47281786 | 3.83 |
ENSRNOT00000089123
|
Kcnj12
|
potassium voltage-gated channel subfamily J member 12 |
| chr14_+_2789650 | 3.81 |
ENSRNOT00000030479
|
Fam69a
|
family with sequence similarity 69, member A |
| chr2_-_164684985 | 3.77 |
ENSRNOT00000057504
|
Rarres1
|
retinoic acid receptor responder 1 |
| chr1_+_86923591 | 3.69 |
ENSRNOT00000027166
|
Sars2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr15_-_19733967 | 3.59 |
ENSRNOT00000012036
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr3_+_125503638 | 3.50 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr8_-_1450138 | 3.46 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr3_-_22951711 | 3.39 |
ENSRNOT00000016876
ENSRNOT00000085981 |
Psmb7
|
proteasome subunit beta 7 |
| chr13_+_103300932 | 3.34 |
ENSRNOT00000085214
ENSRNOT00000038264 |
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr1_+_88686731 | 3.26 |
ENSRNOT00000028222
|
Polr2i
|
RNA polymerase II subunit I |
| chr1_-_86923575 | 3.25 |
ENSRNOT00000027029
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr10_+_109614965 | 3.21 |
ENSRNOT00000071432
|
Tspan10
|
tetraspanin 10 |
| chr10_-_56531483 | 3.18 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_209768696 | 3.14 |
ENSRNOT00000022406
|
Glrx3
|
glutaredoxin 3 |
| chr14_+_60764409 | 3.11 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr12_+_47218969 | 3.05 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr10_+_43633035 | 3.04 |
ENSRNOT00000077764
|
Igtp
|
interferon gamma induced GTPase |
| chr5_-_152446775 | 3.01 |
ENSRNOT00000021919
|
Catsper4
|
cation channel, sperm associated 4 |
| chr8_-_53146953 | 3.01 |
ENSRNOT00000045356
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr1_+_86948918 | 3.00 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr10_+_86711240 | 2.97 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr2_+_189400696 | 2.96 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr8_+_59278262 | 2.96 |
ENSRNOT00000017053
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr5_+_63056089 | 2.94 |
ENSRNOT00000081090
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
| chr1_+_85112834 | 2.91 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr2_-_210550490 | 2.90 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr18_+_72005581 | 2.88 |
ENSRNOT00000072519
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr7_-_117259791 | 2.87 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr2_-_197814808 | 2.85 |
ENSRNOT00000074156
|
Adamtsl4
|
ADAMTS-like 4 |
| chr4_+_116968000 | 2.84 |
ENSRNOT00000020786
|
Emx1
|
empty spiracles homeobox 1 |
| chr7_+_126939925 | 2.82 |
ENSRNOT00000022746
|
Gramd4
|
GRAM domain containing 4 |
| chr15_-_34198921 | 2.80 |
ENSRNOT00000024991
|
Nrl
|
neural retina leucine zipper |
| chr3_+_56030915 | 2.71 |
ENSRNOT00000010489
|
Phospho2
|
phosphatase, orphan 2 |
| chr19_+_54553419 | 2.69 |
ENSRNOT00000025392
|
Jph3
|
junctophilin 3 |
| chr15_+_51756978 | 2.68 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr1_-_168144280 | 2.61 |
ENSRNOT00000020969
|
Olr69
|
olfactory receptor 69 |
| chr4_+_32373641 | 2.60 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr15_-_34898237 | 2.59 |
ENSRNOT00000091428
|
AABR07017936.1
|
|
| chr3_-_22952063 | 2.55 |
ENSRNOT00000083850
|
Psmb7
|
proteasome subunit beta 7 |
| chr6_-_138931429 | 2.54 |
ENSRNOT00000090584
|
LOC100359993
|
Ighg protein-like |
| chr7_-_144272578 | 2.53 |
ENSRNOT00000020676
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr5_+_134737030 | 2.47 |
ENSRNOT00000066161
ENSRNOT00000057061 |
Kncn
|
kinocilin |
| chr17_-_89923990 | 2.46 |
ENSRNOT00000071150
|
LOC100910957
|
acyl-CoA-binding domain-containing protein 5-like |
| chr1_+_213334956 | 2.45 |
ENSRNOT00000073278
|
Olr309
|
olfactory receptor 309 |
| chr11_+_70034139 | 2.43 |
ENSRNOT00000002450
|
Umps
|
uridine monophosphate synthetase |
| chr10_+_34631398 | 2.40 |
ENSRNOT00000046027
|
Olr1391
|
olfactory receptor 1391 |
| chr10_+_14668316 | 2.33 |
ENSRNOT00000091891
|
LOC685321
|
similar to protease, serine, 28 |
| chr6_-_141062581 | 2.30 |
ENSRNOT00000073446
|
AABR07065778.3
|
|
| chr17_-_89923423 | 2.28 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr2_+_168228099 | 2.23 |
ENSRNOT00000074681
|
Vom1r56
|
vomeronasal 1 receptor 56 |
| chr4_-_122237754 | 2.15 |
ENSRNOT00000029915
|
Chst13
|
carbohydrate sulfotransferase 13 |
| chr9_-_113331319 | 2.08 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr13_-_93746994 | 2.05 |
ENSRNOT00000005072
|
Opn3
|
opsin 3 |
| chr13_-_88307988 | 2.04 |
ENSRNOT00000003812
|
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr11_+_27364916 | 2.04 |
ENSRNOT00000002151
|
Bach1
|
BTB domain and CNC homolog 1 |
| chr17_-_23923792 | 1.98 |
ENSRNOT00000018739
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr17_+_85356042 | 1.88 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr1_-_125967756 | 1.87 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr2_-_128002005 | 1.85 |
ENSRNOT00000018796
|
Pgrmc2
|
progesterone receptor membrane component 2 |
| chr10_+_34402482 | 1.84 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr9_+_94310921 | 1.82 |
ENSRNOT00000026646
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr6_+_93539271 | 1.78 |
ENSRNOT00000078791
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr12_-_13998172 | 1.72 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr4_+_119997268 | 1.70 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr1_-_212633304 | 1.70 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chr16_+_20668971 | 1.70 |
ENSRNOT00000027073
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr2_-_179704629 | 1.64 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_+_9578345 | 1.64 |
ENSRNOT00000021756
|
Mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr5_-_163167299 | 1.59 |
ENSRNOT00000022478
|
Tnfrsf1b
|
TNF receptor superfamily member 1B |
| chr4_+_342302 | 1.58 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr19_-_39646693 | 1.58 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr3_+_65815080 | 1.58 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_-_168725656 | 1.57 |
ENSRNOT00000040224
|
Olr115
|
olfactory receptor 115 |
| chr12_-_24578761 | 1.51 |
ENSRNOT00000076209
|
Tbl2
|
transducin (beta)-like 2 |
| chr3_+_154507035 | 1.48 |
ENSRNOT00000017265
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr8_-_25669394 | 1.48 |
ENSRNOT00000036738
|
Dpy19l1
|
dpy-19 like 1 |
| chr1_-_171028743 | 1.47 |
ENSRNOT00000026488
|
Olr223
|
olfactory receptor 223 |
| chr8_-_115981910 | 1.45 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr1_+_82452469 | 1.44 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr7_-_2986935 | 1.42 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr9_+_100080144 | 1.42 |
ENSRNOT00000071411
|
Rnpepl1
|
arginyl aminopeptidase like 1 |
| chr5_-_28349927 | 1.42 |
ENSRNOT00000008725
|
Otud6b
|
OTU domain containing 6B |
| chr1_-_167308827 | 1.41 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr7_+_130474279 | 1.41 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr9_+_10013854 | 1.40 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr7_-_136853957 | 1.38 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr20_-_5533448 | 1.35 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr10_-_45923829 | 1.35 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
| chr17_+_23986758 | 1.34 |
ENSRNOT00000024066
|
Sirt5
|
sirtuin 5 |
| chr3_-_51297852 | 1.34 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr10_-_79097807 | 1.32 |
ENSRNOT00000003421
|
Kif2b
|
kinesin family member 2B |
| chr6_-_28464118 | 1.32 |
ENSRNOT00000068214
|
Efr3b
|
EFR3 homolog B |
| chr12_-_12782139 | 1.29 |
ENSRNOT00000001392
ENSRNOT00000079836 |
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr9_-_66033871 | 1.28 |
ENSRNOT00000035209
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr5_+_63050758 | 1.27 |
ENSRNOT00000009452
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
| chr1_-_214074663 | 1.27 |
ENSRNOT00000026215
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr1_+_218487824 | 1.26 |
ENSRNOT00000017958
|
Mrgprd
|
MAS related GPR family member D |
| chr8_+_5734348 | 1.25 |
ENSRNOT00000013119
|
Mmp10
|
matrix metallopeptidase 10 |
| chrX_+_156716604 | 1.23 |
ENSRNOT00000092207
|
Irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_-_18058055 | 1.22 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr3_+_112371677 | 1.21 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr4_+_84854386 | 1.21 |
ENSRNOT00000013620
|
Mturn
|
maturin, neural progenitor differentiation regulator homolog |
| chr5_+_154205402 | 1.19 |
ENSRNOT00000064925
|
Srsf10
|
serine and arginine rich splicing factor 10 |
| chr7_-_75364166 | 1.18 |
ENSRNOT00000036852
|
Snx31
|
sorting nexin 31 |
| chr5_-_154363329 | 1.18 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chr1_+_154377447 | 1.15 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_+_42852683 | 1.15 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr3_-_165700489 | 1.10 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr3_-_176791960 | 1.10 |
ENSRNOT00000018237
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr7_-_139929895 | 1.10 |
ENSRNOT00000047272
|
Olr1104
|
olfactory receptor 1104 |
| chr3_-_175479395 | 1.07 |
ENSRNOT00000077308
|
Hrh3
|
histamine receptor H3 |
| chr9_+_94310469 | 1.06 |
ENSRNOT00000065743
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr3_-_79165698 | 1.02 |
ENSRNOT00000032617
|
Olr744
|
olfactory receptor 744 |
| chr10_-_6870011 | 1.00 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr10_+_88764732 | 0.98 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chrX_+_42558931 | 0.98 |
ENSRNOT00000044129
|
Ptchd1
|
patched domain containing 1 |
| chr20_-_5533600 | 0.97 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr15_+_4658246 | 0.96 |
ENSRNOT00000008391
|
Saysd1
|
SAYSVFN motif domain containing 1 |
| chr20_+_7818289 | 0.96 |
ENSRNOT00000042539
|
Ppard
|
peroxisome proliferator-activated receptor delta |
| chr3_-_94419048 | 0.95 |
ENSRNOT00000015775
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr1_+_214534284 | 0.94 |
ENSRNOT00000064254
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr3_+_138504214 | 0.93 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
| chr1_+_128199322 | 0.89 |
ENSRNOT00000075131
|
Lysmd4
|
LysM domain containing 4 |
| chr10_+_102136283 | 0.88 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr13_-_47670407 | 0.87 |
ENSRNOT00000035044
|
Il19
|
interleukin 19 |
| chr2_-_187863503 | 0.85 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr9_+_29984077 | 0.85 |
ENSRNOT00000075592
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 |
| chr4_-_121969446 | 0.81 |
ENSRNOT00000085001
|
Vom1r99
|
vomeronasal 1 receptor 99 |
| chr13_-_88061108 | 0.80 |
ENSRNOT00000003774
|
Rgs4
|
regulator of G-protein signaling 4 |
| chr14_-_2934571 | 0.79 |
ENSRNOT00000049772
|
Ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr10_-_31052102 | 0.78 |
ENSRNOT00000007917
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr6_-_42473738 | 0.77 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr7_+_12043794 | 0.74 |
ENSRNOT00000039813
|
Atp8b3
|
ATPase phospholipid transporting 8B3 |
| chr8_-_55050194 | 0.73 |
ENSRNOT00000013197
|
LOC100151767
|
hypothetical LOC100151767 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 2.9 | 8.6 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 2.7 | 13.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 2.7 | 8.0 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 2.6 | 7.7 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 2.5 | 9.9 | GO:1904313 | response to methamphetamine hydrochloride(GO:1904313) |
| 2.4 | 9.7 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 2.3 | 9.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 2.3 | 6.8 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) hydrogen sulfide biosynthetic process(GO:0070814) |
| 2.0 | 6.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 1.8 | 5.4 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 1.7 | 5.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.5 | 4.6 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.5 | 10.5 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 1.4 | 4.2 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.4 | 4.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 1.3 | 11.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 1.2 | 13.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 1.2 | 4.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.1 | 3.3 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 1.0 | 5.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.0 | 3.0 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.0 | 2.9 | GO:1904124 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.9 | 10.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.9 | 2.8 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.9 | 4.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.8 | 9.1 | GO:1903874 | ferrous iron transport(GO:0015684) zinc II ion transmembrane import(GO:0071578) ferrous iron transmembrane transport(GO:1903874) |
| 0.8 | 4.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.8 | 12.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.8 | 5.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 11.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.7 | 3.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.6 | 3.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.6 | 3.0 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.6 | 3.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.6 | 2.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 4.8 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.5 | 3.2 | GO:0045901 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.5 | 5.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.5 | 7.3 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.5 | 4.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.5 | 3.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.5 | 1.4 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.5 | 1.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.5 | 6.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 1.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 1.2 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.4 | 8.0 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.4 | 1.5 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 2.8 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 2.4 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 1.3 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.3 | 1.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 1.4 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.3 | 3.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 2.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 3.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.3 | 2.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 5.9 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 4.6 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.2 | 1.4 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 4.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 0.7 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.2 | 2.0 | GO:1990637 | estrogen biosynthetic process(GO:0006703) response to prolactin(GO:1990637) |
| 0.2 | 2.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 2.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 1.6 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 0.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 1.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 1.3 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.2 | 1.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.2 | 1.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.2 | 1.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.2 | 1.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 7.0 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.2 | 8.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 8.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.2 | 4.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 2.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 2.0 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 6.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 2.7 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 10.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 3.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.3 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 3.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 3.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 3.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 4.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 4.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.6 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 3.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.9 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.1 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.3 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.1 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 2.9 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 2.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 5.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.4 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.0 | 1.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 3.8 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.7 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 14.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 1.3 | 2.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.2 | 8.6 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 1.1 | 4.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.0 | 11.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 1.0 | 5.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.0 | 2.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 3.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 8.0 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.6 | 7.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.5 | 3.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.5 | 1.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 4.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 3.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 26.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 3.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 1.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.4 | 3.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 3.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 5.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 2.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 4.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 2.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 2.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 1.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 3.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.9 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.2 | 2.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 4.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 4.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 11.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 7.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 13.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 6.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 3.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 5.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 9.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 7.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 5.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 2.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 5.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 3.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 4.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 7.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 12.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 5.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 16.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 2.6 | 7.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.8 | 7.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.8 | 7.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.7 | 13.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.5 | 4.6 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.5 | 10.5 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 1.5 | 11.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.3 | 9.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.1 | 9.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 1.1 | 3.3 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 1.0 | 9.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 1.0 | 3.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.0 | 6.8 | GO:1904047 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) S-adenosyl-L-methionine binding(GO:1904047) |
| 1.0 | 2.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.8 | 6.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.7 | 2.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.6 | 6.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.6 | 3.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 12.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 5.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.5 | 2.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.5 | 4.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 5.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 2.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 8.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 3.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 5.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 4.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 5.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 7.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.3 | 9.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 3.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 11.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 4.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.2 | 2.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 5.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 4.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.8 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 3.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 2.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 5.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 4.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 2.7 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 5.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 3.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 5.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 4.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 10.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 2.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 5.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 3.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 2.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 3.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 3.6 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.4 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 11.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 6.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 14.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 6.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 4.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 10.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.2 | 4.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 10.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 14.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.5 | 8.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 8.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 9.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 6.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 3.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 2.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 6.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 8.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 3.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 1.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 9.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 12.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 3.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 1.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 5.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 3.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 4.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 2.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 3.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 4.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 6.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 3.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |