Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hdx
Z-value: 0.85
Transcription factors associated with Hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hdx
|
ENSRNOG00000037799 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | rn6_v1_chrX_-_82986051_82986051 | -0.10 | 6.7e-02 | Click! |
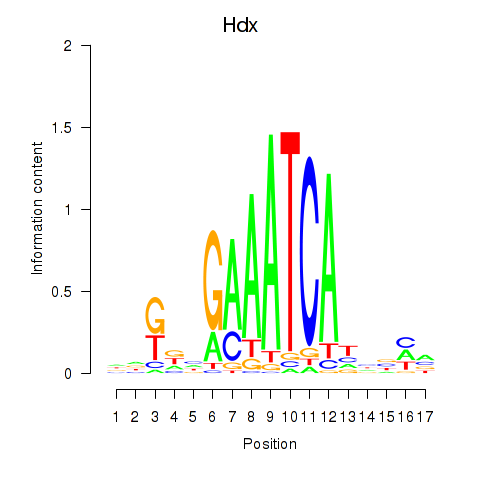
Activity profile of Hdx motif
Sorted Z-values of Hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_8489266 | 20.12 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_-_28799266 | 19.06 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr14_+_13751231 | 18.63 |
ENSRNOT00000044244
|
Gk2
|
glycerol kinase 2 |
| chr9_+_23503236 | 17.28 |
ENSRNOT00000017996
|
Crisp2
|
cysteine-rich secretory protein 2 |
| chrX_-_32153794 | 17.21 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr9_+_4817854 | 16.60 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr2_+_200785507 | 16.11 |
ENSRNOT00000046942
|
Hao2
|
hydroxyacid oxidase 2 |
| chr4_-_128266082 | 15.77 |
ENSRNOT00000039549
|
Nup50
|
nucleoporin 50 |
| chr1_-_54036068 | 15.12 |
ENSRNOT00000075019
|
RGD1560718
|
similar to putative protein kinase |
| chr16_+_68633720 | 14.83 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr4_-_176231344 | 13.00 |
ENSRNOT00000049154
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chrX_+_62282212 | 12.88 |
ENSRNOT00000039568
|
AABR07038837.1
|
|
| chr10_+_71202456 | 12.70 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr14_-_21748356 | 12.69 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr8_+_85503224 | 12.43 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr1_-_76517134 | 12.22 |
ENSRNOT00000064593
ENSRNOT00000085775 |
LOC100912485
|
alcohol sulfotransferase-like |
| chr3_+_104749051 | 11.78 |
ENSRNOT00000000010
|
Tmco5b
|
transmembrane and coiled-coil domains 5B |
| chr1_-_54854353 | 11.59 |
ENSRNOT00000072895
|
Smok2a
|
sperm motility kinase 2A |
| chrX_-_158261717 | 11.53 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr3_+_143119687 | 11.34 |
ENSRNOT00000006608
|
Cst12
|
cystatin 12 |
| chr7_-_70643169 | 11.13 |
ENSRNOT00000010106
|
Inhbe
|
inhibin beta E subunit |
| chr17_-_43537293 | 10.99 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chrX_-_143558521 | 10.64 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr18_-_53181503 | 10.53 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr6_+_101288951 | 10.30 |
ENSRNOT00000046901
|
RGD1562540
|
RGD1562540 |
| chr4_+_56711275 | 9.95 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr14_+_22142364 | 9.83 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chrX_-_144001727 | 9.72 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr1_-_266428239 | 9.63 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr1_-_67302751 | 9.51 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr18_-_19149470 | 9.45 |
ENSRNOT00000061146
|
Spata45
|
spermatogenesis associated 45 |
| chr5_-_77433847 | 9.38 |
ENSRNOT00000076906
ENSRNOT00000043056 |
LOC500473
Mup4
|
similar to alpha-2u globulin PGCL2 major urinary protein 4 |
| chr17_-_66667853 | 8.98 |
ENSRNOT00000058749
|
A26c2
|
ANKRD26-like family C, member 2 |
| chr4_-_163095614 | 8.94 |
ENSRNOT00000088759
|
RGD1564770
|
similar to CD69 antigen (p60, early T-cell activation antigen) |
| chrX_-_43781586 | 8.88 |
ENSRNOT00000051551
|
Cldn34b
|
claudin 34B |
| chr3_-_151548995 | 8.72 |
ENSRNOT00000071825
|
LOC102550306
|
uncharacterized LOC102550306 |
| chrX_-_63999622 | 8.67 |
ENSRNOT00000090902
|
LOC103694487
|
protein gar2-like |
| chr17_-_80807181 | 8.51 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr1_+_248723397 | 8.33 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr10_+_96661696 | 8.28 |
ENSRNOT00000077567
|
AABR07030630.1
|
|
| chr5_+_124300477 | 8.11 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr11_-_75144903 | 8.10 |
ENSRNOT00000002329
|
Hrasls
|
HRAS-like suppressor |
| chr1_+_24900858 | 8.07 |
ENSRNOT00000075616
|
LOC501467
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr5_-_110706656 | 7.99 |
ENSRNOT00000033781
|
Izumo3
|
IZUMO family member 3 |
| chr8_-_60359440 | 7.95 |
ENSRNOT00000037353
|
Nxpe2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr20_+_13827132 | 7.85 |
ENSRNOT00000001664
|
Ddt
|
D-dopachrome tautomerase |
| chr5_-_77903062 | 7.72 |
ENSRNOT00000073954
ENSRNOT00000059320 ENSRNOT00000089382 ENSRNOT00000076074 ENSRNOT00000074349 |
LOC100912565
Rn50_5_0814.4
|
major urinary protein-like alpha2u globulin (LOC298111), mRNA |
| chr10_-_87248572 | 7.60 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr16_-_49003246 | 7.58 |
ENSRNOT00000089501
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chrX_-_73562046 | 7.57 |
ENSRNOT00000080501
|
Gm14597
|
predicted gene 14597 |
| chr11_+_80742467 | 7.53 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_-_18531210 | 7.33 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr1_-_67284864 | 7.27 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr20_+_41266566 | 7.26 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr9_+_95161157 | 7.03 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_-_77408323 | 7.00 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chrX_-_98095663 | 6.96 |
ENSRNOT00000004491
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr2_+_22910236 | 6.94 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr6_+_27241919 | 6.84 |
ENSRNOT00000013123
|
Cib4
|
calcium and integrin binding family member 4 |
| chr11_+_44089797 | 6.70 |
ENSRNOT00000060861
|
LOC108352322
|
ferritin heavy chain-like |
| chr19_+_27835890 | 6.58 |
ENSRNOT00000042150
|
LOC100909409
|
disks large homolog 5-like |
| chr5_-_113621134 | 6.55 |
ENSRNOT00000037206
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr7_+_3630950 | 6.53 |
ENSRNOT00000074535
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr10_-_60126696 | 6.42 |
ENSRNOT00000026857
|
Olr1485
|
olfactory receptor 1485 |
| chr9_-_92530938 | 6.35 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr19_-_345698 | 6.32 |
ENSRNOT00000044037
|
LOC501297
|
hypothetical LOC501297 |
| chr1_-_228538074 | 6.13 |
ENSRNOT00000028604
|
Olr324
|
olfactory receptor 324 |
| chr9_+_17911944 | 6.11 |
ENSRNOT00000080940
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chrX_-_152367861 | 6.05 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chrX_+_144994139 | 6.00 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr2_+_104020955 | 5.94 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr10_-_56403188 | 5.91 |
ENSRNOT00000019947
|
Chrnb1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr10_+_64952119 | 5.87 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr3_-_64095120 | 5.86 |
ENSRNOT00000016837
|
Sestd1
|
SEC14 and spectrin domain containing 1 |
| chr7_-_20070772 | 5.84 |
ENSRNOT00000071008
|
LOC102551633
|
sperm motility kinase W-like |
| chr3_-_75235439 | 5.75 |
ENSRNOT00000046688
|
Olr552
|
olfactory receptor 552 |
| chr3_-_103745236 | 5.58 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr12_+_18679789 | 5.57 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr18_+_30592794 | 5.44 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr5_-_156105867 | 5.35 |
ENSRNOT00000078851
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chrX_-_37705263 | 5.34 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr3_-_75089064 | 5.33 |
ENSRNOT00000046167
|
Olr542
|
olfactory receptor 542 |
| chr19_-_28296640 | 5.32 |
ENSRNOT00000041115
ENSRNOT00000042781 |
AABR07043429.1
|
|
| chr2_-_194875962 | 5.28 |
ENSRNOT00000085431
|
RGD1566337
|
similar to TDPOZ2 |
| chr1_-_173469699 | 5.24 |
ENSRNOT00000040345
|
Olr281
|
olfactory receptor 281 |
| chr19_-_462559 | 5.17 |
ENSRNOT00000046676
|
AABR07042633.1
|
|
| chr7_+_144623555 | 5.14 |
ENSRNOT00000022217
|
Hoxc6
|
homeo box C6 |
| chr1_+_83626639 | 4.93 |
ENSRNOT00000028261
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr16_-_86582984 | 4.88 |
ENSRNOT00000057349
|
RGD1564807
|
similar to zinc finger protein, subfamily 1A, 5 |
| chrX_+_25016401 | 4.74 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr6_-_135829953 | 4.73 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr19_+_12097632 | 4.72 |
ENSRNOT00000049452
|
Ces5a
|
carboxylesterase 5A |
| chr4_-_119148358 | 4.72 |
ENSRNOT00000012218
|
Gkn1
|
gastrokine 1 |
| chr2_+_27905535 | 4.69 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr7_+_28414350 | 4.68 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr4_+_162774109 | 4.64 |
ENSRNOT00000084289
|
Klrb1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr8_+_128792707 | 4.60 |
ENSRNOT00000083329
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr1_-_89358166 | 4.56 |
ENSRNOT00000044532
|
Mag
|
myelin-associated glycoprotein |
| chr4_+_154423209 | 4.51 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr10_-_15381691 | 4.43 |
ENSRNOT00000027439
|
Rab11fip3
|
RAB11 family interacting protein 3 |
| chr19_-_22194740 | 4.43 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr8_-_16486419 | 4.40 |
ENSRNOT00000033223
|
LOC690235
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase-like 1) |
| chr3_-_14571640 | 4.34 |
ENSRNOT00000060013
|
Ggta1l1
|
glycoprotein, alpha-galactosyltransferase 1-like 1 |
| chr18_-_63825408 | 4.32 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_76930631 | 4.31 |
ENSRNOT00000032109
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr5_+_29622281 | 4.24 |
ENSRNOT00000012377
|
Nbn
|
nibrin |
| chr10_+_44330897 | 4.24 |
ENSRNOT00000075040
|
LOC100911398
|
olfactory receptor 2AK2-like |
| chr7_-_97067864 | 4.23 |
ENSRNOT00000078009
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr10_+_86399827 | 4.21 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr7_-_107203897 | 4.13 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr15_-_32925673 | 4.05 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr14_-_39112600 | 4.02 |
ENSRNOT00000003170
|
Gabrb1
|
gamma-aminobutyric acid type A receptor beta 1 subunit |
| chr2_-_227145185 | 3.96 |
ENSRNOT00000083189
|
AABR07013202.1
|
|
| chr8_-_28044876 | 3.90 |
ENSRNOT00000072152
|
Acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr15_-_49467174 | 3.79 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr14_+_1318157 | 3.75 |
ENSRNOT00000061889
|
Vom2r67
|
vomeronasal 2 receptor, 67 |
| chr15_+_48674380 | 3.74 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr7_-_124172979 | 3.73 |
ENSRNOT00000074960
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr6_+_48452369 | 3.70 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr13_-_110678389 | 3.69 |
ENSRNOT00000082906
|
AABR07022168.1
|
|
| chr17_+_45670284 | 3.64 |
ENSRNOT00000086536
|
Olr1660
|
olfactory receptor 1660 |
| chr12_-_35979193 | 3.57 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr1_+_168136959 | 3.56 |
ENSRNOT00000020953
|
Olr68
|
olfactory receptor 68 |
| chr3_-_73879647 | 3.55 |
ENSRNOT00000090000
|
Olr510
|
olfactory receptor 510 |
| chr1_-_167588630 | 3.53 |
ENSRNOT00000050707
|
Olr39
|
olfactory receptor 39 |
| chr5_+_69214948 | 3.52 |
ENSRNOT00000074807
|
Olr844
|
olfactory receptor 844 |
| chr4_+_84423653 | 3.52 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr10_-_98469799 | 3.51 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr7_-_141624972 | 3.49 |
ENSRNOT00000078785
|
AABR07058884.1
|
|
| chr8_-_105462141 | 3.38 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr1_+_171188133 | 3.35 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr11_-_71135493 | 3.35 |
ENSRNOT00000050535
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr3_-_113438801 | 3.33 |
ENSRNOT00000091665
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr19_-_49510901 | 3.24 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr5_-_133959447 | 3.22 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr3_+_77921705 | 3.22 |
ENSRNOT00000064006
|
AC132028.1
|
|
| chr3_-_173249556 | 3.10 |
ENSRNOT00000081623
|
AABR07054895.1
|
|
| chr7_-_140770647 | 3.06 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr2_+_115074344 | 3.02 |
ENSRNOT00000073657
|
LOC100910500
|
olfactory receptor 144-like |
| chr1_-_228505080 | 3.01 |
ENSRNOT00000028600
|
Olr322
|
olfactory receptor 322 |
| chr8_+_115179893 | 3.01 |
ENSRNOT00000017469
|
Rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr8_+_58347736 | 2.97 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr15_+_35504845 | 2.96 |
ENSRNOT00000075687
|
Olr1297
|
olfactory receptor 1297 |
| chr7_-_7753978 | 2.96 |
ENSRNOT00000048223
|
Olr1029
|
olfactory receptor 1029 |
| chr15_+_35909279 | 2.94 |
ENSRNOT00000071711
|
Olr1279
|
olfactory receptor 1279 |
| chr6_-_80102635 | 2.94 |
ENSRNOT00000084611
|
Sec23a
|
Sec23 homolog A, coat complex II component |
| chr8_+_73593310 | 2.92 |
ENSRNOT00000012048
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr1_-_103426467 | 2.91 |
ENSRNOT00000045792
|
Mrgprb4
|
MAS-related GPR, member B4 |
| chr8_-_53277859 | 2.88 |
ENSRNOT00000009574
|
Htr3b
|
5-hydroxytryptamine receptor 3B |
| chr15_+_35889080 | 2.83 |
ENSRNOT00000075355
|
LOC100910640
|
olfactory receptor 144-like |
| chr10_+_44407475 | 2.82 |
ENSRNOT00000071556
|
Olr1437
|
olfactory receptor 1437 |
| chr5_-_152227677 | 2.80 |
ENSRNOT00000020051
|
Dhdds
|
dehydrodolichyl diphosphate synthase subunit |
| chr15_+_35813468 | 2.78 |
ENSRNOT00000083638
|
Olr1283
|
olfactory receptor 1283 |
| chr7_-_50278842 | 2.75 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr1_+_169115981 | 2.72 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr3_-_102484517 | 2.71 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chr11_-_80936524 | 2.70 |
ENSRNOT00000044591
|
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr1_-_75849658 | 2.64 |
ENSRNOT00000071105
|
Bsph2
|
binder of sperm protein homolog 2 |
| chr2_-_89310946 | 2.64 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr1_-_149770781 | 2.64 |
ENSRNOT00000082185
|
Olr16
|
olfactory receptor 16 |
| chr1_-_66775384 | 2.60 |
ENSRNOT00000039088
|
Vom1r52
|
vomeronasal 1 receptor 52 |
| chr1_+_173012247 | 2.60 |
ENSRNOT00000073864
|
LOC100912217
|
olfactory receptor 10A3-like |
| chr3_-_78134947 | 2.60 |
ENSRNOT00000079940
|
LOC687065
|
similar to olfactory receptor Olr695 |
| chr1_-_227149748 | 2.59 |
ENSRNOT00000063879
|
Ms4a8
|
membrane spanning 4-domains A8 |
| chrX_+_23414354 | 2.58 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr3_-_151258580 | 2.57 |
ENSRNOT00000026120
|
Edem2
|
ER degradation enhancing alpha-mannosidase like protein 2 |
| chr15_+_19195666 | 2.56 |
ENSRNOT00000041790
|
Ptgdrl
|
prostaglandin D2 receptor-like |
| chr3_+_16183322 | 2.55 |
ENSRNOT00000072630
|
Olr406
|
olfactory receptor 406 |
| chr9_-_82336806 | 2.55 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr2_+_219598162 | 2.52 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr3_+_147226004 | 2.52 |
ENSRNOT00000012765
|
Tmem74b
|
transmembrane protein 74B |
| chr3_+_102456938 | 2.52 |
ENSRNOT00000051827
|
Olr748
|
olfactory receptor 748 |
| chr9_+_118842787 | 2.52 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr3_+_75299283 | 2.49 |
ENSRNOT00000013328
|
Olr555
|
olfactory receptor 555 |
| chr1_-_168703107 | 2.49 |
ENSRNOT00000021334
|
Olr113
|
olfactory receptor 113 |
| chr8_-_37126809 | 2.42 |
ENSRNOT00000040783
|
LOC103693040
|
prostate and testis expressed protein 2-like |
| chr2_-_26438790 | 2.42 |
ENSRNOT00000035017
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr17_+_38457783 | 2.42 |
ENSRNOT00000059757
|
Prl5a2
|
prolactin family 5, subfamily a, member 2 |
| chr7_-_117712213 | 2.41 |
ENSRNOT00000092930
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr1_+_66959610 | 2.39 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr8_+_43781921 | 2.36 |
ENSRNOT00000078003
|
Olr1329
|
olfactory receptor 1329 |
| chr3_+_16039249 | 2.36 |
ENSRNOT00000046196
|
Olr403
|
olfactory receptor 403 |
| chr1_+_258074860 | 2.33 |
ENSRNOT00000054729
|
Cyp2c24
|
cytochrome P450, family 2, subfamily c, polypeptide 24 |
| chr5_+_98998163 | 2.32 |
ENSRNOT00000075600
|
Pramef5
|
PRAME family member 5 |
| chrX_+_105011489 | 2.30 |
ENSRNOT00000085068
|
Arl13a
|
ADP ribosylation factor like GTPase 13A |
| chr13_+_83073544 | 2.29 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chr7_+_36826360 | 2.25 |
ENSRNOT00000029783
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr3_+_55461420 | 2.17 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr10_-_87261717 | 2.17 |
ENSRNOT00000015740
|
Krt27
|
keratin 27 |
| chr1_-_155955173 | 2.17 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr1_-_173393390 | 2.15 |
ENSRNOT00000050657
|
Olr285
|
olfactory receptor 285 |
| chr20_-_10013190 | 2.11 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr3_+_57286892 | 2.10 |
ENSRNOT00000039065
|
LOC103690018
|
Golgi reassembly-stacking protein 2-like |
| chr7_+_6068010 | 2.06 |
ENSRNOT00000044587
|
Olr1007
|
olfactory receptor 1007 |
| chr15_+_35571103 | 2.05 |
ENSRNOT00000085376
|
Olr1293
|
olfactory receptor 1293 |
| chr4_-_13565838 | 2.04 |
ENSRNOT00000048882
|
Olr987
|
olfactory receptor 987 |
| chr11_+_45888221 | 2.00 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr5_+_13379772 | 2.00 |
ENSRNOT00000010032
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr3_-_74991168 | 2.00 |
ENSRNOT00000049273
|
Olr550
|
olfactory receptor 550 |
| chr18_+_30474947 | 1.98 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 5.4 | 16.1 | GO:0018924 | mandelate metabolic process(GO:0018924) |
| 4.2 | 12.7 | GO:0061215 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 3.2 | 9.6 | GO:0010034 | response to acetate(GO:0010034) |
| 2.1 | 10.5 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 1.9 | 17.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.8 | 36.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 1.8 | 5.4 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.7 | 8.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.7 | 26.4 | GO:0051923 | sulfation(GO:0051923) |
| 1.5 | 5.9 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 1.4 | 5.6 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 1.2 | 4.7 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 1.1 | 15.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.1 | 4.2 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 1.0 | 11.0 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 5.9 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.9 | 2.8 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.9 | 4.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.9 | 2.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.9 | 2.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.8 | 4.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.8 | 7.0 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 12.4 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.8 | 7.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) response to oleic acid(GO:0034201) |
| 0.7 | 8.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 8.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.6 | 1.9 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.5 | 1.6 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.5 | 6.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 4.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 4.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.5 | 1.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.5 | 5.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.4 | 6.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 0.9 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.4 | 7.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.4 | 4.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.4 | 1.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 1.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 2.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 4.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 1.9 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.3 | 1.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 4.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 11.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 2.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 3.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 2.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 4.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.4 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 3.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.9 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.2 | 2.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 9.9 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 1.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.4 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.1 | 6.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 7.3 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.9 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.1 | 20.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 1.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.7 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 4.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 5.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 15.7 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 2.6 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 7.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 4.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 133.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 0.9 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 3.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.5 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 10.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.2 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 1.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 2.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 11.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 5.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 3.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 2.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 3.1 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 5.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 2.7 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 2.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 4.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 36.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.5 | 4.6 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 1.3 | 13.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.2 | 8.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.9 | 4.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.8 | 8.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.7 | 5.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.6 | 8.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.6 | 4.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 4.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.5 | 2.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 16.9 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 3.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 2.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.3 | 2.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 5.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 12.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 4.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 29.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 0.8 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 13.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.5 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 2.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 5.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 5.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 4.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 4.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 8.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 7.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 7.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 6.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.7 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 26.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 5.2 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 69.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 5.4 | 16.1 | GO:0052853 | very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 3.6 | 14.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 3.3 | 16.4 | GO:0005550 | pheromone binding(GO:0005550) |
| 2.6 | 7.8 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 2.4 | 36.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 1.9 | 7.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.5 | 5.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.4 | 9.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.4 | 6.9 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.2 | 8.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.2 | 3.5 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 1.1 | 3.3 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 1.1 | 11.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 1.1 | 4.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.9 | 2.6 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.7 | 12.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.7 | 2.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 5.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 9.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.6 | 4.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.6 | 2.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 4.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.5 | 4.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 15.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 3.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 4.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.4 | 2.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 10.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 4.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 5.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.3 | 6.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 13.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 7.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 4.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 3.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 11.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 0.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 16.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 4.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 3.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 2.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 13.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 7.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 4.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.9 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.5 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.2 | 1.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 10.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 2.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.2 | 11.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 4.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 7.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 131.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 12.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 8.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 13.0 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.1 | 6.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 1.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 11.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 4.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 6.7 | GO:0004672 | protein kinase activity(GO:0004672) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 12.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 9.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 18.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 4.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 4.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 12.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 6.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.1 | 9.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.8 | 7.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.8 | 9.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 10.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 7.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 8.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 12.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.4 | 7.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 12.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 9.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 9.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 2.6 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 6.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 3.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 2.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 4.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 1.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |