Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
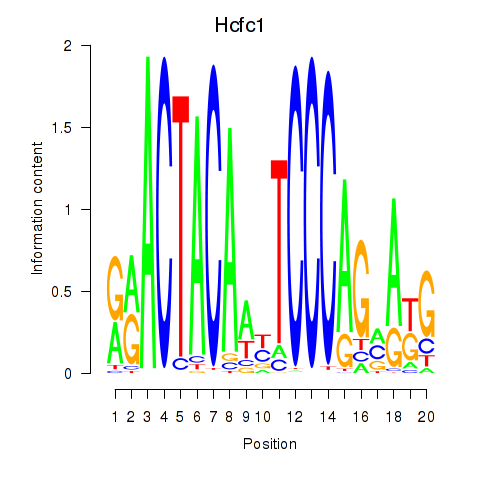
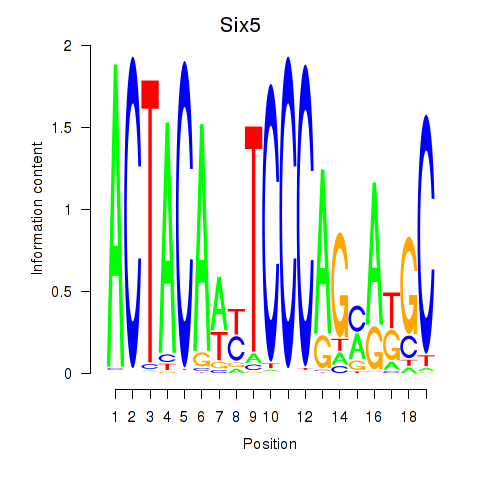
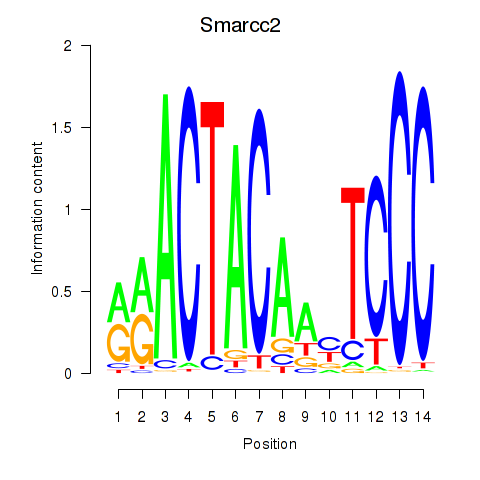
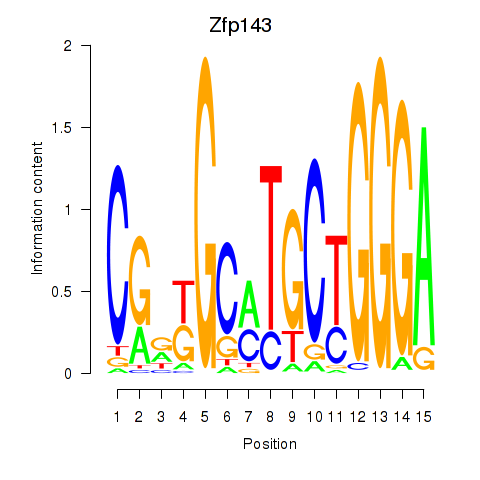
Results for Hcfc1_Six5_Smarcc2_Zfp143
Z-value: 2.43
Transcription factors associated with Hcfc1_Six5_Smarcc2_Zfp143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hcfc1
|
ENSRNOG00000051948 | host cell factor C1 |
|
Six5
|
ENSRNOG00000060146 | SIX homeobox 5 |
|
Smarcc2
|
ENSRNOG00000031135 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
Zfp143
|
ENSRNOG00000010087 | zinc finger protein 143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hcfc1 | rn6_v1_chrX_+_156812064_156812064 | 0.38 | 3.8e-12 | Click! |
| Zfp143 | rn6_v1_chr1_+_174702373_174702373 | 0.26 | 1.7e-06 | Click! |
| Smarcc2 | rn6_v1_chr7_+_2875909_2875909 | 0.24 | 1.0e-05 | Click! |
| Six5 | rn6_v1_chr1_+_80000165_80000165 | -0.04 | 4.6e-01 | Click! |
Activity profile of Hcfc1_Six5_Smarcc2_Zfp143 motif
Sorted Z-values of Hcfc1_Six5_Smarcc2_Zfp143 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_143063983 | 37.15 |
ENSRNOT00000006329
|
Napb
|
NSF attachment protein beta |
| chr4_-_61573003 | 32.83 |
ENSRNOT00000012179
|
Slc35b4
|
solute carrier family 35 member B4 |
| chr14_-_80169431 | 31.32 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr15_-_4454958 | 29.91 |
ENSRNOT00000070933
|
Nudt13
|
nudix hydrolase 13 |
| chr1_-_226255886 | 28.30 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr7_+_117417687 | 27.28 |
ENSRNOT00000018240
|
Maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr11_+_84396033 | 26.67 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr7_-_12429897 | 23.80 |
ENSRNOT00000020670
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr6_+_99883959 | 23.09 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr16_+_36116258 | 22.40 |
ENSRNOT00000017652
|
Sap30
|
Sin3A associated protein 30 |
| chr5_+_165922915 | 21.83 |
ENSRNOT00000089179
ENSRNOT00000059522 |
Dffa
|
DNA fragmentation factor subunit alpha |
| chr11_+_47146308 | 21.79 |
ENSRNOT00000002191
|
Cep97
|
centrosomal protein 97 |
| chr2_-_41785792 | 21.09 |
ENSRNOT00000015871
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr2_-_187258140 | 20.86 |
ENSRNOT00000017735
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr5_+_135663371 | 19.79 |
ENSRNOT00000024375
|
Mutyh
|
mutY DNA glycosylase |
| chr7_-_145450233 | 19.69 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr10_+_98706960 | 19.67 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr4_+_118655728 | 19.36 |
ENSRNOT00000043082
|
Aak1
|
AP2 associated kinase 1 |
| chr1_+_220307394 | 19.36 |
ENSRNOT00000071891
|
LOC108348052
|
equilibrative nucleoside transporter 2 |
| chr2_+_188495629 | 18.99 |
ENSRNOT00000027828
|
Fam189b
|
family with sequence similarity 189, member B |
| chr14_-_84937725 | 18.96 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_-_82108083 | 18.75 |
ENSRNOT00000027677
ENSRNOT00000084530 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr7_-_144322240 | 18.72 |
ENSRNOT00000089290
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr12_+_7454884 | 18.42 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr1_+_220400855 | 18.35 |
ENSRNOT00000027178
|
Slc29a2
|
solute carrier family 29 member 2 |
| chr12_+_23789638 | 17.97 |
ENSRNOT00000001954
ENSRNOT00000084730 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr4_+_7282948 | 17.84 |
ENSRNOT00000011052
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr3_+_148327965 | 17.74 |
ENSRNOT00000010851
ENSRNOT00000088481 |
Tpx2
|
TPX2, microtubule nucleation factor |
| chr6_+_129835919 | 17.50 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr8_+_28045288 | 17.45 |
ENSRNOT00000085997
|
Thyn1
|
thymocyte nuclear protein 1 |
| chr5_+_173542358 | 17.39 |
ENSRNOT00000027347
|
RGD1311517
|
similar to RIKEN cDNA 9430015G10 |
| chr3_+_8363937 | 17.35 |
ENSRNOT00000019500
|
Slc27a4
|
solute carrier family 27 member 4 |
| chr1_+_81373340 | 17.23 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr4_+_28441301 | 17.10 |
ENSRNOT00000067087
|
Vps50
|
VPS50 EARP/GARPII complex subunit |
| chr3_+_110367939 | 16.71 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr10_-_12994495 | 16.60 |
ENSRNOT00000004672
|
Thoc6
|
THO complex 6 |
| chr2_+_174013288 | 16.42 |
ENSRNOT00000013904
|
Serpini1
|
serpin family I member 1 |
| chr3_-_55951584 | 16.31 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr8_+_28045093 | 16.23 |
ENSRNOT00000073373
|
Thyn1
|
thymocyte nuclear protein 1 |
| chr7_+_11660934 | 16.12 |
ENSRNOT00000022336
|
Lmnb2
|
lamin B2 |
| chr1_+_75356220 | 15.95 |
ENSRNOT00000019799
|
Lig1
|
DNA ligase 1 |
| chr14_-_106393670 | 15.61 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chr9_-_93607087 | 15.55 |
ENSRNOT00000025262
|
Pde6d
|
phosphodiesterase 6D |
| chr5_+_135562034 | 15.50 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr18_-_81807627 | 14.99 |
ENSRNOT00000020376
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr9_+_60883981 | 14.98 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr12_-_25638797 | 14.82 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr18_-_28495937 | 14.74 |
ENSRNOT00000027194
|
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr10_+_16259728 | 14.73 |
ENSRNOT00000028115
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr20_-_46666830 | 14.61 |
ENSRNOT00000000331
|
Cep57l1
|
centrosomal protein 57-like 1 |
| chr5_-_135561914 | 14.43 |
ENSRNOT00000023178
|
Mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr5_+_78267248 | 14.29 |
ENSRNOT00000019910
|
Prpf4
|
pre-mRNA processing factor 4 |
| chr4_-_7311807 | 14.28 |
ENSRNOT00000012222
ENSRNOT00000089817 |
Abcb8
|
ATP binding cassette subfamily B member 8 |
| chr20_+_18547370 | 14.08 |
ENSRNOT00000000750
|
Ube2d1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr7_-_83348487 | 13.94 |
ENSRNOT00000006685
|
Nudcd1
|
NudC domain containing 1 |
| chr4_-_60358562 | 13.93 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr11_+_88831130 | 13.48 |
ENSRNOT00000002478
|
Dnm1l
|
dynamin 1-like |
| chr14_+_81725513 | 13.40 |
ENSRNOT00000020154
|
Zfyve28
|
zinc finger FYVE-type containing 28 |
| chr10_-_51669297 | 13.37 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr11_+_88830957 | 13.32 |
ENSRNOT00000002482
ENSRNOT00000002477 |
Dnm1l
|
dynamin 1-like |
| chr10_+_10967658 | 13.23 |
ENSRNOT00000004999
|
Cdip1
|
cell death-inducing p53 target 1 |
| chr2_+_93518806 | 13.20 |
ENSRNOT00000050987
|
Snx16
|
sorting nexin 16 |
| chr4_+_123118468 | 12.99 |
ENSRNOT00000010895
|
Tmem43
|
transmembrane protein 43 |
| chr2_+_187893368 | 12.81 |
ENSRNOT00000092031
|
Mex3a
|
mex-3 RNA binding family member A |
| chr20_-_48503898 | 12.77 |
ENSRNOT00000073091
|
Wasf1
|
WAS protein family, member 1 |
| chr3_+_11114551 | 12.73 |
ENSRNOT00000013507
|
Plpp7
|
phospholipid phosphatase 7 |
| chr1_+_100473643 | 12.52 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr11_+_32450587 | 12.47 |
ENSRNOT00000061235
|
Smim11
|
small integral membrane protein 11 |
| chr8_-_62332115 | 12.19 |
ENSRNOT00000025783
|
Mpi
|
mannose phosphate isomerase (mapped) |
| chr9_+_93607477 | 12.14 |
ENSRNOT00000025411
|
Cops7b
|
COP9 signalosome subunit 7B |
| chr2_+_54897424 | 12.02 |
ENSRNOT00000017665
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr4_-_157347803 | 11.92 |
ENSRNOT00000020785
|
Usp5
|
ubiquitin specific peptidase 5 |
| chr1_+_142679345 | 11.71 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr1_-_266142538 | 11.68 |
ENSRNOT00000026792
|
Actr1a
|
ARP1 actin-related protein 1 homolog A, centractin alpha |
| chr5_+_152681101 | 11.57 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr1_-_84491466 | 11.56 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr3_-_177078594 | 11.54 |
ENSRNOT00000020695
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr7_+_143897014 | 11.53 |
ENSRNOT00000017314
|
Espl1
|
extra spindle pole bodies like 1, separase |
| chr19_-_22632071 | 11.39 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr4_-_119568736 | 11.01 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr4_-_123118186 | 10.98 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr2_-_211001258 | 10.72 |
ENSRNOT00000037336
|
Atxn7l2
|
ataxin 7-like 2 |
| chr8_-_61823102 | 10.60 |
ENSRNOT00000058645
|
Neil1
|
nei-like DNA glycosylase 1 |
| chr1_-_63293635 | 10.59 |
ENSRNOT00000090337
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr3_+_110918243 | 10.50 |
ENSRNOT00000056432
|
Rad51
|
RAD51 recombinase |
| chr2_-_58534211 | 10.48 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr6_+_26546924 | 10.48 |
ENSRNOT00000007763
|
Eif2b4
|
eukaryotic translation initiation factor 2B subunit delta |
| chr17_-_54070399 | 10.43 |
ENSRNOT00000022924
|
B3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr5_-_1347921 | 10.42 |
ENSRNOT00000007994
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1 |
| chr1_-_71399751 | 10.31 |
ENSRNOT00000020787
|
Zfp444
|
zinc finger protein 444 |
| chr20_+_45458558 | 10.17 |
ENSRNOT00000000713
|
Cdk19
|
cyclin-dependent kinase 19 |
| chr19_+_54766589 | 10.15 |
ENSRNOT00000025894
|
Banp
|
Btg3 associated nuclear protein |
| chr5_-_136748980 | 10.09 |
ENSRNOT00000026844
|
Ipo13
|
importin 13 |
| chr8_+_116297950 | 10.01 |
ENSRNOT00000051522
|
Nprl2
|
NPR2-like, GATOR1 complex subunit |
| chr5_-_138222534 | 9.97 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr11_+_61748883 | 9.97 |
ENSRNOT00000093552
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr10_-_104179523 | 9.87 |
ENSRNOT00000005292
|
Slc25a19
|
solute carrier family 25 member 19 |
| chr17_-_54070231 | 9.84 |
ENSRNOT00000085150
|
B3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr16_+_81587446 | 9.80 |
ENSRNOT00000092680
|
NEWGENE_1582994
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr10_+_39655455 | 9.70 |
ENSRNOT00000058817
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_+_187288644 | 9.65 |
ENSRNOT00000035271
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chr4_+_57050214 | 9.64 |
ENSRNOT00000025165
|
Ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr7_-_2986935 | 9.63 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr10_-_49196177 | 9.52 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr1_-_68269117 | 9.52 |
ENSRNOT00000072367
|
LOC100911224
|
zinc finger protein interacting with ribonucleoprotein K-like |
| chr1_+_157403595 | 9.45 |
ENSRNOT00000012781
|
Ccdc90b
|
coiled-coil domain containing 90B |
| chr7_-_36597462 | 9.38 |
ENSRNOT00000066322
|
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr2_-_198184739 | 9.37 |
ENSRNOT00000028751
|
Vps45
|
vacuolar protein sorting 45 |
| chr9_+_16612433 | 9.37 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr7_-_97902585 | 9.31 |
ENSRNOT00000008382
|
RGD1310852
|
similar to RIKEN cDNA 9130401M01 |
| chr15_-_41770112 | 9.24 |
ENSRNOT00000034400
|
Kpna3
|
karyopherin subunit alpha 3 |
| chr7_-_34951644 | 9.20 |
ENSRNOT00000030015
|
Vezt
|
vezatin, adherens junctions transmembrane protein |
| chr9_-_66877972 | 9.19 |
ENSRNOT00000023463
|
Wdr12
|
WD repeat domain 12 |
| chr1_-_220136470 | 9.19 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr3_-_91195981 | 9.16 |
ENSRNOT00000056935
|
RGD1309730
|
similar to RIKEN cDNA B230118H07 |
| chr13_+_68785827 | 8.87 |
ENSRNOT00000003517
|
Trmt1l
|
tRNA methyltransferase 1-like |
| chr4_-_121565212 | 8.86 |
ENSRNOT00000088753
|
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr7_-_140342727 | 8.85 |
ENSRNOT00000089612
|
Ddx23
|
DEAD-box helicase 23 |
| chr7_-_142132173 | 8.85 |
ENSRNOT00000026613
|
Csrnp2
|
cysteine and serine rich nuclear protein 2 |
| chr10_+_38877422 | 8.80 |
ENSRNOT00000065229
ENSRNOT00000083162 |
Sept8
|
septin 8 |
| chr3_+_57817693 | 8.80 |
ENSRNOT00000067729
ENSRNOT00000047243 ENSRNOT00000013400 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr1_-_64175099 | 8.73 |
ENSRNOT00000091288
|
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr9_-_10801140 | 8.62 |
ENSRNOT00000072771
|
Fem1a
|
fem-1 homolog A |
| chr1_+_81372650 | 8.59 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr1_-_13395370 | 8.58 |
ENSRNOT00000085519
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr1_-_71400388 | 8.56 |
ENSRNOT00000090327
|
Zfp444
|
zinc finger protein 444 |
| chr3_+_177188044 | 8.51 |
ENSRNOT00000022073
|
Tcea2
|
transcription elongation factor A2 |
| chr3_-_3476215 | 8.51 |
ENSRNOT00000024352
|
Ubac1
|
UBA domain containing 1 |
| chr2_-_196304169 | 8.49 |
ENSRNOT00000028639
|
Scnm1
|
sodium channel modifier 1 |
| chr7_-_2786856 | 8.44 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chr5_+_101526551 | 8.43 |
ENSRNOT00000015254
|
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr20_+_3422461 | 8.41 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr10_-_86645529 | 8.40 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr8_+_53365857 | 8.29 |
ENSRNOT00000080712
|
Zw10
|
zw10 kinetochore protein |
| chr2_-_189880743 | 8.27 |
ENSRNOT00000018830
|
Snapin
|
SNAP-associated protein |
| chr5_-_136053210 | 8.20 |
ENSRNOT00000025903
|
Kif2c
|
kinesin family member 2C |
| chr3_-_11382004 | 8.20 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chr7_+_97778260 | 8.19 |
ENSRNOT00000064264
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr12_-_23727535 | 8.17 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr1_-_86948845 | 8.17 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr12_-_47438931 | 8.13 |
ENSRNOT00000050782
|
RGD1311899
|
similar to RIKEN cDNA 2210016L21 gene |
| chr5_-_78267063 | 8.13 |
ENSRNOT00000040894
ENSRNOT00000087648 |
Cdc26
|
cell division cycle 26 |
| chr1_-_100865894 | 8.09 |
ENSRNOT00000027580
|
Ptov1
|
prostate tumor overexpressed 1 |
| chr9_+_19880473 | 8.02 |
ENSRNOT00000052068
|
Slc25a27
|
solute carrier family 25, member 27 |
| chr10_+_83476107 | 8.01 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr10_-_15346236 | 8.00 |
ENSRNOT00000088104
ENSRNOT00000027430 |
Capn15
|
calpain 15 |
| chr19_-_38321528 | 7.94 |
ENSRNOT00000031977
|
Smpd3
|
sphingomyelin phosphodiesterase 3 |
| chr3_-_150885475 | 7.93 |
ENSRNOT00000034708
ENSRNOT00000038809 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr3_-_111013589 | 7.92 |
ENSRNOT00000016692
|
Dnajc17
|
DnaJ heat shock protein family (Hsp40) member C17 |
| chr1_+_203526850 | 7.89 |
ENSRNOT00000087065
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr10_-_36336628 | 7.86 |
ENSRNOT00000030035
|
Zfp879
|
zinc finger protein 879 |
| chr6_+_36107147 | 7.83 |
ENSRNOT00000006020
|
Rdh14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr7_+_2435553 | 7.83 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr10_+_15538872 | 7.82 |
ENSRNOT00000060078
|
Luc7l
|
LUC7-like |
| chr10_+_46940965 | 7.78 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr14_+_106393959 | 7.78 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chrX_+_71155601 | 7.77 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr4_+_7122890 | 7.76 |
ENSRNOT00000076339
ENSRNOT00000076418 ENSRNOT00000014718 |
Abcf2
|
ATP binding cassette subfamily F member 2 |
| chr19_+_38039564 | 7.74 |
ENSRNOT00000087491
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chrX_-_64908682 | 7.70 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr7_+_25919867 | 7.68 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chrX_+_15742978 | 7.67 |
ENSRNOT00000015546
|
Ccdc22
|
coiled-coil domain containing 22 |
| chr13_-_51784639 | 7.60 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_40315377 | 7.52 |
ENSRNOT00000091319
ENSRNOT00000036141 |
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr16_-_54137660 | 7.46 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chr4_+_78024765 | 7.42 |
ENSRNOT00000043856
|
Krba1
|
KRAB-A domain containing 1 |
| chr5_+_126976456 | 7.36 |
ENSRNOT00000014275
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr5_-_135663284 | 7.35 |
ENSRNOT00000087237
ENSRNOT00000066869 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr3_+_35014538 | 7.33 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chr9_+_79944132 | 7.30 |
ENSRNOT00000071856
ENSRNOT00000022459 |
Smarcal1
|
Swi/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr1_+_87790104 | 7.29 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr8_-_28044876 | 7.29 |
ENSRNOT00000072152
|
Acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chrX_-_15620841 | 7.28 |
ENSRNOT00000085397
|
Praf2
|
PRA1 domain family, member 2 |
| chr8_-_106617597 | 7.27 |
ENSRNOT00000070953
|
Mrps22
|
mitochondrial ribosomal protein S22 |
| chr16_-_54137485 | 7.24 |
ENSRNOT00000014202
|
Pcm1
|
pericentriolar material 1 |
| chr5_+_50002184 | 7.21 |
ENSRNOT00000010962
|
Akirin2
|
akirin 2 |
| chr7_-_29152442 | 7.16 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr10_-_106883423 | 7.14 |
ENSRNOT00000071406
|
LOC688286
|
similar to threonine aldolase 1 |
| chr1_-_185569190 | 7.11 |
ENSRNOT00000090773
|
RGD1311703
|
similar to sid2057p |
| chr11_+_60907015 | 7.10 |
ENSRNOT00000002797
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr18_+_70996044 | 7.09 |
ENSRNOT00000024950
ENSRNOT00000066336 |
Dym
|
dymeclin |
| chr10_+_12994683 | 7.09 |
ENSRNOT00000004702
|
Hcfc1r1
|
host cell factor C1 regulator 1 |
| chr6_-_26546545 | 7.02 |
ENSRNOT00000091264
|
Snx17
|
sorting nexin 17 |
| chr12_+_2201891 | 7.01 |
ENSRNOT00000091197
ENSRNOT00000001325 |
Retn
|
resistin |
| chr4_-_112707270 | 6.99 |
ENSRNOT00000009308
|
Mrpl19
|
mitochondrial ribosomal protein L19 |
| chr2_+_206392200 | 6.99 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chr6_+_111296417 | 6.97 |
ENSRNOT00000075425
|
Ahsa1
|
activator of Hsp90 ATPase activity 1 |
| chr1_+_87248489 | 6.95 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr2_+_62150251 | 6.94 |
ENSRNOT00000016196
|
Zfr
|
zinc finger RNA binding protein |
| chr6_-_111176918 | 6.94 |
ENSRNOT00000016242
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chr1_-_100530183 | 6.85 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr14_+_81340041 | 6.80 |
ENSRNOT00000082612
ENSRNOT00000066391 |
Nop14
|
NOP14 nucleolar protein |
| chrX_-_84821775 | 6.78 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr19_+_37830917 | 6.75 |
ENSRNOT00000025608
|
Nutf2
|
nuclear transport factor 2 |
| chr8_+_126411829 | 6.71 |
ENSRNOT00000013450
|
Azi2
|
5-azacytidine induced 2 |
| chr1_+_199351628 | 6.70 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr1_+_226947105 | 6.69 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr5_+_2042991 | 6.66 |
ENSRNOT00000050236
|
Eloc
|
elongin C |
| chr1_-_197801634 | 6.66 |
ENSRNOT00000090200
|
Nfatc2ip
|
nuclear factor of activated T-cells 2 interacting protein |
| chr18_-_76984203 | 6.65 |
ENSRNOT00000089122
|
Ctdp1
|
CTD phosphatase subunit 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hcfc1_Six5_Smarcc2_Zfp143
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.8 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 9.3 | 37.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 9.1 | 27.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 8.9 | 26.8 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 7.7 | 23.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 7.4 | 44.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 6.1 | 18.3 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 5.4 | 21.5 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 4.9 | 19.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 4.7 | 18.8 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 4.6 | 32.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 4.4 | 17.5 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 3.7 | 11.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 3.4 | 17.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 3.3 | 13.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 3.3 | 9.9 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 3.2 | 9.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 3.1 | 9.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 3.0 | 15.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 2.9 | 17.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.9 | 11.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 2.8 | 8.4 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 2.7 | 11.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 2.6 | 18.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 2.4 | 7.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 2.4 | 12.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 2.4 | 7.1 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 2.3 | 7.0 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 2.3 | 7.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 2.3 | 6.9 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 2.3 | 20.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 2.2 | 6.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 2.2 | 24.6 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 2.2 | 15.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 2.2 | 17.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 2.1 | 8.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 2.1 | 6.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 2.1 | 6.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 2.0 | 14.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 2.0 | 8.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 2.0 | 8.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 2.0 | 6.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 2.0 | 23.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 2.0 | 5.9 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 2.0 | 5.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 1.9 | 5.8 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 1.9 | 11.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.9 | 7.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.9 | 44.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 1.8 | 7.3 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 1.8 | 10.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.7 | 7.0 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 1.7 | 12.2 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 1.7 | 18.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 1.7 | 19.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 1.6 | 6.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 1.6 | 4.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.6 | 6.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.6 | 4.8 | GO:0042245 | RNA repair(GO:0042245) |
| 1.6 | 6.3 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 1.5 | 1.5 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.5 | 4.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.5 | 11.8 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 1.5 | 8.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 1.4 | 2.9 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.4 | 18.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.4 | 9.9 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 1.4 | 14.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.4 | 2.8 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 1.4 | 4.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.4 | 8.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.3 | 10.8 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 1.3 | 4.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.3 | 21.3 | GO:0042407 | cristae formation(GO:0042407) |
| 1.3 | 6.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.3 | 15.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 1.3 | 7.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.3 | 1.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 1.3 | 15.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.3 | 6.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.3 | 12.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.3 | 5.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.3 | 16.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 1.2 | 2.5 | GO:1905146 | regulation of protein catabolic process in the vacuole(GO:1904350) lysosomal protein catabolic process(GO:1905146) |
| 1.2 | 14.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.2 | 8.2 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.1 | 5.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.1 | 5.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.1 | 4.4 | GO:0051958 | methotrexate transport(GO:0051958) |
| 1.1 | 10.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.1 | 3.2 | GO:2001034 | histone H3-K36 dimethylation(GO:0097676) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.1 | 6.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.1 | 6.3 | GO:0009838 | abscission(GO:0009838) |
| 1.0 | 4.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 1.0 | 4.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.0 | 25.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.0 | 7.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.0 | 2.9 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.9 | 2.8 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.9 | 1.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.9 | 3.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.9 | 6.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.9 | 2.7 | GO:0006431 | methionyl-tRNA aminoacylation(GO:0006431) |
| 0.9 | 14.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.9 | 2.6 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.9 | 1.7 | GO:0061740 | late endosomal microautophagy(GO:0061738) protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.9 | 12.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.9 | 11.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.8 | 6.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.8 | 2.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.8 | 10.9 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.8 | 10.0 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.8 | 5.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.8 | 4.2 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.8 | 13.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.8 | 19.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.8 | 3.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.8 | 6.3 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.8 | 3.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.8 | 3.1 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.8 | 6.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.8 | 3.1 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.7 | 1.5 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.7 | 4.4 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.7 | 8.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.7 | 2.2 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.7 | 2.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.7 | 5.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.7 | 3.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.7 | 2.1 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.7 | 2.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.7 | 2.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.7 | 2.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.7 | 17.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.7 | 2.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.6 | 7.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.6 | 1.9 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.6 | 5.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.6 | 5.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 | 4.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.6 | 6.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.6 | 1.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.6 | 5.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.6 | 1.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.6 | 9.6 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.6 | 1.2 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.6 | 4.2 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.6 | 4.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.6 | 1.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.6 | 6.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.6 | 12.7 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.6 | 1.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.6 | 2.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.6 | 1.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.6 | 1.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.6 | 4.0 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.6 | 2.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.6 | 8.9 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.6 | 4.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.5 | 4.4 | GO:0046959 | habituation(GO:0046959) |
| 0.5 | 2.7 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.5 | 2.2 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.5 | 2.7 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.5 | 1.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.5 | 2.1 | GO:0015746 | citrate transport(GO:0015746) |
| 0.5 | 3.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 10.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 1.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 6.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.5 | 2.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.5 | 11.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.5 | 6.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.5 | 10.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.5 | 3.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 1.5 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.5 | 4.9 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.5 | 1.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.5 | 6.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.5 | 4.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.5 | 11.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.5 | 1.4 | GO:0050883 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.5 | 6.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 8.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.5 | 3.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.5 | 14.9 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.5 | 3.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 3.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.4 | 4.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 15.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 3.5 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 6.5 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.4 | 3.9 | GO:2000344 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 1.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.4 | 5.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.4 | 4.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 2.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.4 | 9.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.4 | 2.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.4 | 1.2 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.4 | 4.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 3.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 3.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.4 | 1.2 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.4 | 2.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 3.9 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.4 | 5.0 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.4 | 2.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.4 | 14.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.4 | 1.5 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 3.4 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.4 | 10.4 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.4 | 3.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.4 | 4.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.4 | 1.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 18.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.4 | 2.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.4 | 23.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.4 | 2.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.4 | 1.4 | GO:0097681 | double-strand break repair via alternative nonhomologous end joining(GO:0097681) |
| 0.3 | 2.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 | 1.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.3 | 6.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.3 | 1.4 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.3 | 1.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.3 | 7.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.3 | 22.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.3 | 1.7 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.3 | 5.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 7.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 19.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.3 | 13.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.3 | 2.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 2.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 28.4 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.3 | 11.1 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.3 | 0.9 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.3 | 17.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.3 | 1.2 | GO:0070203 | regulation of establishment of protein localization to telomere(GO:0070203) positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.3 | 5.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 0.6 | GO:0000019 | regulation of mitotic recombination(GO:0000019) negative regulation of telomere capping(GO:1904354) |
| 0.3 | 2.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.3 | 0.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 3.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 3.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 0.6 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 1.7 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.3 | 1.9 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 2.4 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.3 | 3.3 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 1.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.3 | 1.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 1.3 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.3 | 7.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.3 | 40.4 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.3 | 1.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.2 | 2.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.2 | 0.7 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.2 | 1.7 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.2 | 1.0 | GO:1904796 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.2 | 8.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 1.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 0.9 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 2.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.2 | 0.9 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.2 | 3.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 9.4 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.2 | 1.8 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 2.5 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 1.1 | GO:0034982 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.2 | 5.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.2 | 1.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 5.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 0.9 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.2 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 2.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 2.5 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.2 | 7.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.2 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.8 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 0.8 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 24.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.2 | 3.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 14.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 3.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 19.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.2 | 2.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.5 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.2 | 8.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 | 5.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 1.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 2.6 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.2 | 5.1 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.2 | 3.9 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) |
| 0.2 | 1.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.2 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 3.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 0.5 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 2.9 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 11.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 10.9 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 1.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.6 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 1.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 10.8 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 6.9 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 1.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 1.0 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.9 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 1.3 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.1 | 4.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 2.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 2.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 3.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 6.5 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.1 | 3.8 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.9 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.6 | GO:0046477 | galactosylceramide metabolic process(GO:0006681) glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 11.9 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.1 | 4.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 1.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 3.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 5.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 2.9 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.1 | 1.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 2.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.1 | 7.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 28.6 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.1 | 0.7 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 5.8 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 0.5 | GO:0002176 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.1 | 2.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 5.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 2.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 6.7 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.1 | 8.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 8.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 1.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 14.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 6.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.1 | 7.8 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 13.9 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 1.0 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 2.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) |
| 0.1 | 3.7 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) |
| 0.1 | 4.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 2.0 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.1 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.8 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 1.7 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 122.0 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.1 | 2.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 5.5 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.1 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 3.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 15.2 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 6.9 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.0 | GO:0071307 | cellular response to vitamin K(GO:0071307) |
| 0.0 | 0.3 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 1.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.8 | GO:0016573 | histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 2.9 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 2.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 37.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 5.9 | 17.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 4.8 | 14.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 4.5 | 17.8 | GO:1990745 | EARP complex(GO:1990745) |
| 3.6 | 10.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.1 | 24.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 2.9 | 23.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 2.7 | 8.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 2.6 | 10.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.5 | 10.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 2.4 | 17.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 2.3 | 28.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 2.2 | 17.7 | GO:0043203 | axon hillock(GO:0043203) |
| 2.1 | 10.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.1 | 6.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 2.0 | 6.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 2.0 | 6.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 2.0 | 16.1 | GO:0005638 | lamin filament(GO:0005638) |
| 2.0 | 10.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 2.0 | 19.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.9 | 19.3 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 1.8 | 5.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.8 | 5.3 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 1.7 | 10.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.7 | 8.7 | GO:0030689 | Noc complex(GO:0030689) |
| 1.7 | 6.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.6 | 14.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.6 | 8.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.6 | 18.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.5 | 9.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 1.5 | 15.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 1.5 | 8.8 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 1.5 | 8.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 1.4 | 11.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.3 | 6.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.3 | 11.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.3 | 18.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.3 | 2.6 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.3 | 6.5 | GO:0097361 | CIA complex(GO:0097361) |
| 1.3 | 3.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.2 | 7.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.1 | 7.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 5.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 1.1 | 10.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.1 | 40.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 1.0 | 2.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.0 | 6.3 | GO:0089701 | U2AF(GO:0089701) |
| 1.0 | 5.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.0 | 10.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.9 | 23.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.9 | 13.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.9 | 6.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.9 | 6.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.9 | 12.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.9 | 8.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.9 | 1.7 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.9 | 4.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.8 | 4.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.8 | 39.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.8 | 8.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.8 | 2.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.8 | 48.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.8 | 4.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.8 | 5.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 6.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.8 | 6.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.7 | 8.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.7 | 2.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.7 | 13.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.7 | 2.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.7 | 2.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.7 | 2.8 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.7 | 2.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.7 | 5.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.7 | 4.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.7 | 22.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.7 | 8.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.6 | 7.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.6 | 8.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.6 | 8.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.6 | 28.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.6 | 2.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.6 | 22.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 6.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.5 | 5.4 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.5 | 2.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.5 | 25.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.5 | 9.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.5 | 6.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 2.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 5.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.5 | 1.5 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.5 | 6.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.5 | 4.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.5 | 2.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 6.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.5 | 12.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 49.4 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.4 | 3.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 8.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 7.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.4 | 5.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.4 | 2.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 2.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 2.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 8.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 10.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.4 | 11.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 2.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 16.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 5.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 10.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 2.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.4 | 18.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 1.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 12.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 5.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 5.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 22.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 2.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 4.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 20.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 0.6 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.3 | 4.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 25.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 2.3 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.3 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 12.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.3 | 5.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.3 | 5.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 11.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 2.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.3 | 1.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 5.3 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 11.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 2.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 8.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 16.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 9.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.2 | 55.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 3.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.8 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.2 | 13.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 2.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 18.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.2 | 12.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 2.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 1.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 7.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 2.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 3.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.6 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 31.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 16.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 55.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 1.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 37.6 | GO:0030016 | myofibril(GO:0030016) |
| 0.2 | 12.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 21.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.2 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 5.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 4.0 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.1 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 9.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 4.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 24.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 26.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 94.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 2.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 7.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 24.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 17.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 18.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 11.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 5.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 24.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 7.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 8.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 14.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 15.1 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 13.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 41.9 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 9.6 | 9.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 8.2 | 32.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 5.7 | 28.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 5.5 | 27.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 4.2 | 21.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 4.1 | 20.5 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 3.9 | 15.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 3.8 | 23.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 3.8 | 11.4 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 3.6 | 17.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 3.5 | 17.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 3.3 | 9.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 3.0 | 8.9 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 2.9 | 11.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.7 | 8.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 2.5 | 25.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 2.5 | 19.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 2.5 | 19.8 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 2.3 | 11.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 2.2 | 21.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 2.1 | 8.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 2.1 | 6.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 2.1 | 6.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 2.0 | 8.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 2.0 | 6.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 2.0 | 5.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 2.0 | 7.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.9 | 18.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.8 | 29.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.8 | 5.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.8 | 16.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.8 | 29.8 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 1.8 | 10.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.7 | 6.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.7 | 5.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 1.6 | 19.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.6 | 4.9 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 1.6 | 6.4 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.6 | 18.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 1.5 | 7.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.5 | 19.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 1.5 | 29.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.5 | 11.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 1.5 | 8.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 1.4 | 8.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 1.2 | 5.0 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 1.2 | 4.8 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 1.2 | 6.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.2 | 14.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 1.2 | 12.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.2 | 18.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.1 | 8.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.1 | 3.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.1 | 5.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 1.1 | 32.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 1.1 | 7.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.1 | 6.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.1 | 4.4 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 1.1 | 14.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.1 | 3.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.1 | 2.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 1.0 | 14.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 1.0 | 8.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.0 | 4.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.9 | 20.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.9 | 2.7 | GO:0004825 | methionine-tRNA ligase activity(GO:0004825) |
| 0.9 | 6.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.9 | 17.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.9 | 2.6 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.9 | 18.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.9 | 17.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.9 | 4.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.8 | 21.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.8 | 6.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.8 | 4.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.8 | 7.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.8 | 3.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.8 | 4.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.8 | 3.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 3.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.8 | 7.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.7 | 3.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 5.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.7 | 7.3 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.7 | 2.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.7 | 20.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.7 | 10.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.7 | 2.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.7 | 6.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 2.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.7 | 7.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.7 | 2.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.6 | 1.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.6 | 1.9 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.6 | 3.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.6 | 7.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 2.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.6 | 4.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 21.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.6 | 4.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 21.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.6 | 7.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.6 | 19.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 8.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.5 | 2.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.5 | 5.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 2.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.5 | 4.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 26.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.5 | 4.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 6.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 13.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.5 | 1.5 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.5 | 1.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.5 | 7.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.5 | 9.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.5 | 2.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 6.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.5 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 14.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.5 | 1.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 12.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.5 | 15.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.5 | 6.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 8.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 10.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 1.7 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.4 | 8.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.4 | 8.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 4.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 5.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.4 | 1.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.4 | 3.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.4 | 43.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.4 | 3.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 2.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 3.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 2.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 7.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 7.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.3 | 1.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 5.2 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.3 | 2.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 8.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 1.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 2.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 24.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.3 | 2.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.3 | 3.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 1.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 3.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 18.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 0.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 7.5 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.3 | 3.9 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.3 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 9.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 1.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 10.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 0.7 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.2 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 5.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 2.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 7.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 3.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.2 | 10.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 4.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 55.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 6.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 0.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 6.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.2 | 1.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.2 | 5.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 10.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 1.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 5.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 27.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 24.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 4.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 2.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 2.0 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 7.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 15.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) cobalamin binding(GO:0031419) |
| 0.1 | 0.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 4.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.9 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 1.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 16.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 1.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 2.1 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 23.0 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 0.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 24.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.5 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.1 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.6 | GO:0001190 | RNA polymerase II transcription coactivator activity(GO:0001105) transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 12.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.0 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 3.7 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 21.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 76.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 11.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 10.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 1.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 160.4 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.2 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.3 | GO:0034212 | peptide N-acetyltransferase activity(GO:0034212) |
| 0.1 | 1.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 0.4 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 2.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.1 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 1.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 2.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 2.1 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 3.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.3 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 2.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.6 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 5.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0001155 | RNA polymerase III transcription factor binding(GO:0001025) TFIIIA-class transcription factor binding(GO:0001155) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 32.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.4 | 15.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.2 | 21.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 1.0 | 44.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.9 | 34.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.8 | 13.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.7 | 35.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.7 | 22.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.7 | 24.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.6 | 22.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.6 | 26.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 6.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.5 | 32.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.4 | 11.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 11.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 17.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 10.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 6.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 9.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 4.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 6.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 17.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 14.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 5.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 10.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 8.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 2.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 8.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 2.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 6.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 53.7 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 2.5 | 46.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 2.3 | 24.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 2.0 | 31.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.9 | 66.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 1.5 | 23.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 1.4 | 26.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 1.4 | 4.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 1.3 | 6.6 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 1.3 | 2.6 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 1.1 | 19.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.0 | 66.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.9 | 19.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.9 | 8.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.9 | 8.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.9 | 5.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.8 | 14.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.7 | 2.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.7 | 12.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.7 | 18.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.7 | 1.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.6 | 14.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.6 | 51.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.6 | 11.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.6 | 1.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.6 | 3.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.5 | 7.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.5 | 6.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 10.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.5 | 14.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.5 | 61.6 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.4 | 21.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.4 | 15.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 10.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 7.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.4 | 1.2 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.4 | 6.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.4 | 25.7 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.4 | 5.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.4 | 4.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 3.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 8.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 8.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 8.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 6.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 10.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 6.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 4.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 10.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 6.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.3 | 16.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 2.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 2.5 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.2 | 2.7 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.2 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 18.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 8.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 8.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 3.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 4.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 6.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 2.0 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.2 | 4.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 14.1 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.9 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 1.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 9.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 5.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.8 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.1 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 6.3 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 4.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 7.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 4.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |