Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
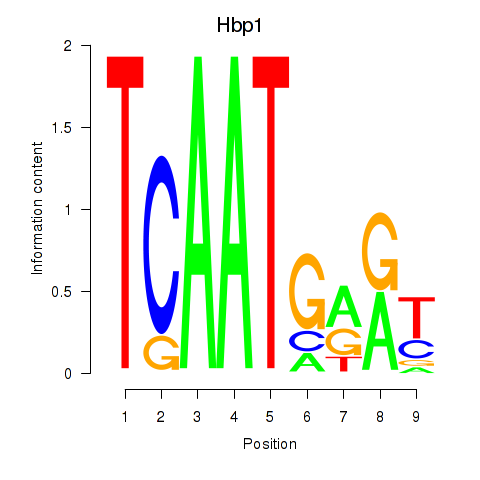
Results for Hbp1
Z-value: 0.89
Transcription factors associated with Hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hbp1
|
ENSRNOG00000008927 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hbp1 | rn6_v1_chr6_-_51257625_51257625 | -0.50 | 4.2e-22 | Click! |
Activity profile of Hbp1 motif
Sorted Z-values of Hbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114423533 | 26.24 |
ENSRNOT00000091221
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr2_+_23289374 | 17.66 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr9_+_4817854 | 17.00 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr4_+_154215250 | 16.16 |
ENSRNOT00000072465
|
Mug2
|
murinoglobulin 2 |
| chr1_-_13915594 | 15.67 |
ENSRNOT00000015927
|
Arfgef3
|
ARFGEF family member 3 |
| chr4_+_65110746 | 15.44 |
ENSRNOT00000017675
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr9_-_26707571 | 15.27 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chrX_+_107496072 | 14.66 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chrX_-_13601069 | 14.61 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr1_+_83714347 | 13.72 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr7_-_129970550 | 13.61 |
ENSRNOT00000055879
|
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr5_-_134207847 | 13.56 |
ENSRNOT00000083686
ENSRNOT00000088209 ENSRNOT00000051252 |
Cyp4a2
|
cytochrome P450, family 4, subfamily a, polypeptide 2 |
| chr1_+_257970345 | 13.42 |
ENSRNOT00000088853
|
Cyp2c11
|
cytochrome P450, subfamily 2, polypeptide 11 |
| chr18_-_35071619 | 13.35 |
ENSRNOT00000075695
|
LOC100911558
|
serine protease inhibitor Kazal-type 3-like |
| chr1_-_258766881 | 13.01 |
ENSRNOT00000015801
|
Cyp2c12
|
cytochrome P450, family 2, subfamily c, polypeptide 12 |
| chr7_-_138483612 | 12.67 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr13_-_91427575 | 12.56 |
ENSRNOT00000012092
|
Apcs
|
amyloid P component, serum |
| chr14_+_22375955 | 12.27 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr9_+_4107246 | 12.26 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr2_+_78247448 | 12.00 |
ENSRNOT00000089805
|
LOC103689968
|
protein FAM134B |
| chr9_+_74124016 | 11.51 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr8_-_85645718 | 11.34 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chr3_-_14112851 | 11.33 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr9_-_4327679 | 10.89 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr9_+_95501778 | 10.87 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr3_-_46726946 | 10.80 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr2_-_235161263 | 10.65 |
ENSRNOT00000080235
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr1_+_248723397 | 10.65 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr4_-_161850875 | 10.42 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr9_-_4945352 | 10.18 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr14_+_22724399 | 10.13 |
ENSRNOT00000002724
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_-_182038178 | 9.80 |
ENSRNOT00000040708
|
Fgb
|
fibrinogen beta chain |
| chr14_+_22724070 | 9.62 |
ENSRNOT00000089471
|
Ugt2b10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr16_-_36161089 | 9.52 |
ENSRNOT00000017888
|
Scrg1
|
stimulator of chondrogenesis 1 |
| chr1_+_67340809 | 9.46 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr17_+_72160735 | 9.46 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_-_132143470 | 9.40 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr9_-_30844199 | 9.27 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr10_-_27862868 | 9.14 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr14_-_46153212 | 9.08 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr18_-_7081356 | 9.04 |
ENSRNOT00000021253
|
Chst9
|
carbohydrate sulfotransferase 9 |
| chr7_+_38858062 | 8.98 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr3_-_52447622 | 8.88 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr10_-_107539658 | 8.78 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr8_-_49109981 | 8.76 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr1_+_55219773 | 8.54 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr11_+_57505005 | 8.52 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr5_-_124403195 | 8.48 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr16_-_32868680 | 8.38 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr12_+_32103198 | 8.26 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr12_-_11733099 | 8.13 |
ENSRNOT00000051244
ENSRNOT00000087257 |
Cyp3a23/3a1
|
cytochrome P450, family 3, subfamily a, polypeptide 23/polypeptide 1 |
| chr16_+_18690246 | 8.05 |
ENSRNOT00000081484
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr17_-_43584152 | 8.02 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr2_+_220298245 | 7.99 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr13_-_39643361 | 7.89 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr1_+_65851060 | 7.87 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr3_-_25212049 | 7.86 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_-_258875572 | 7.86 |
ENSRNOT00000093005
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr8_-_76579099 | 7.84 |
ENSRNOT00000088628
|
Fam81a
|
family with sequence similarity 81, member A |
| chr11_+_28692708 | 7.83 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr7_+_34326087 | 7.82 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr9_+_37727942 | 7.80 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr6_+_127946686 | 7.78 |
ENSRNOT00000082680
|
LOC500712
|
Ab1-233 |
| chr2_+_74360622 | 7.73 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr1_+_83744238 | 7.64 |
ENSRNOT00000028249
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr1_+_72889270 | 7.64 |
ENSRNOT00000058843
ENSRNOT00000034957 |
Tnnt1
|
troponin T1, slow skeletal type |
| chr18_+_35574002 | 7.62 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr1_+_189940291 | 7.60 |
ENSRNOT00000075035
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr1_+_263554453 | 7.54 |
ENSRNOT00000070861
|
Abcc2
|
ATP binding cassette subfamily C member 2 |
| chr1_+_189289957 | 7.50 |
ENSRNOT00000020587
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr14_+_66598259 | 7.49 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr11_+_88732381 | 7.48 |
ENSRNOT00000078367
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_-_2045817 | 7.46 |
ENSRNOT00000009542
ENSRNOT00000081171 ENSRNOT00000078765 |
Gria4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr2_+_55775274 | 7.43 |
ENSRNOT00000018545
|
C9
|
complement C9 |
| chr6_-_127508452 | 7.36 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr8_-_47094352 | 7.35 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr2_+_58448917 | 7.30 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr10_+_5930298 | 7.28 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr7_-_30162056 | 7.24 |
ENSRNOT00000009910
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr13_-_72063347 | 7.15 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr5_-_140657745 | 7.14 |
ENSRNOT00000019080
|
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr3_-_147849875 | 7.13 |
ENSRNOT00000081153
|
Nrsn2
|
neurensin 2 |
| chr13_-_56693968 | 6.91 |
ENSRNOT00000060160
|
AABR07021096.1
|
|
| chr1_+_193187972 | 6.90 |
ENSRNOT00000090172
ENSRNOT00000065342 |
Slc5a11
|
solute carrier family 5 member 11 |
| chr8_+_97291580 | 6.88 |
ENSRNOT00000018794
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr13_+_42220251 | 6.79 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr18_+_29191731 | 6.77 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr20_-_5123073 | 6.70 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr11_+_60072727 | 6.68 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr9_+_6966908 | 6.60 |
ENSRNOT00000072796
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr6_+_29977797 | 6.59 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr14_+_20266891 | 6.58 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr2_-_231648122 | 6.54 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr5_-_147303346 | 6.49 |
ENSRNOT00000009153
|
Hpca
|
hippocalcin |
| chrX_-_142248369 | 6.46 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr10_+_53621375 | 6.43 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr11_+_66316606 | 6.43 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr8_-_76579387 | 6.41 |
ENSRNOT00000090747
|
Fam81a
|
family with sequence similarity 81, member A |
| chr20_-_9855443 | 6.39 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr7_+_20262680 | 6.35 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr2_-_216382244 | 6.34 |
ENSRNOT00000086695
ENSRNOT00000087259 |
LOC103689940
|
pancreatic alpha-amylase-like |
| chr2_+_22909569 | 6.27 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr1_-_112811936 | 6.26 |
ENSRNOT00000093339
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr20_+_41100071 | 6.24 |
ENSRNOT00000000658
|
Tspyl4
|
TSPY-like 4 |
| chr18_+_62852303 | 6.24 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr9_+_95295701 | 6.21 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr17_-_9762813 | 6.19 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr5_+_64294321 | 6.17 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr7_-_71226150 | 6.17 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr2_-_196402106 | 6.15 |
ENSRNOT00000028664
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr2_+_248649441 | 6.11 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr1_+_37507276 | 6.09 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr1_+_167937026 | 6.03 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chr1_-_224698514 | 5.96 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr8_-_84632817 | 5.96 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr12_+_38377034 | 5.93 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr1_-_258877045 | 5.92 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr6_+_128050250 | 5.82 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr8_-_84506328 | 5.81 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_+_145715969 | 5.74 |
ENSRNOT00000037996
|
Tmc3
|
transmembrane channel-like 3 |
| chr1_+_148240504 | 5.74 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr1_-_148119857 | 5.73 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr7_+_20462081 | 5.69 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr18_+_30515962 | 5.69 |
ENSRNOT00000027172
|
LOC108348233
|
protocadherin beta-6-like |
| chr11_-_81660395 | 5.68 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr6_-_108415093 | 5.59 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr9_+_95202632 | 5.56 |
ENSRNOT00000025652
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr5_-_64718131 | 5.52 |
ENSRNOT00000088211
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr10_-_107539465 | 5.51 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr2_-_22744407 | 5.50 |
ENSRNOT00000073710
|
Cmya5
|
cardiomyopathy associated 5 |
| chr17_+_89452814 | 5.50 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr3_-_72219246 | 5.50 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr2_+_147496229 | 5.46 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr14_-_19132208 | 5.46 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chrX_+_28072826 | 5.45 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr1_+_265298868 | 5.43 |
ENSRNOT00000023278
|
Dpcd
|
deleted in primary ciliary dyskinesia |
| chrX_-_13279082 | 5.36 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr3_+_117416345 | 5.34 |
ENSRNOT00000056054
|
Ctxn2
|
cortexin 2 |
| chr8_+_48718329 | 5.31 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chrX_-_29648359 | 5.29 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr18_-_18079560 | 5.28 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr4_-_176528110 | 5.22 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr8_-_73164620 | 5.20 |
ENSRNOT00000031988
|
Tln2
|
talin 2 |
| chr6_-_26445787 | 5.20 |
ENSRNOT00000084242
|
LOC103692570
|
dihydropyrimidinase-related protein 5-like |
| chr10_+_53740841 | 5.19 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr17_+_32082937 | 5.13 |
ENSRNOT00000074775
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr8_-_82641536 | 5.10 |
ENSRNOT00000014491
|
Scg3
|
secretogranin III |
| chr1_+_70260041 | 4.98 |
ENSRNOT00000020416
|
Zim1
|
zinc finger, imprinted 1 |
| chr9_-_66019065 | 4.96 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr2_-_210738378 | 4.95 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr17_+_32973695 | 4.95 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr9_-_75528644 | 4.93 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr3_-_37803112 | 4.92 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr6_+_78567970 | 4.92 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr10_-_96015499 | 4.90 |
ENSRNOT00000004383
|
Cacng4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr4_+_138441332 | 4.89 |
ENSRNOT00000090847
|
Cntn4
|
contactin 4 |
| chr2_-_250805445 | 4.89 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr10_+_71217966 | 4.88 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr18_-_37096132 | 4.87 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr4_+_21462779 | 4.87 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr18_+_52215682 | 4.86 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chrX_-_31759161 | 4.85 |
ENSRNOT00000041515
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr8_+_80965255 | 4.84 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr5_-_93244202 | 4.82 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr9_+_101005082 | 4.82 |
ENSRNOT00000082949
|
Neu4
|
neuraminidase 4 |
| chr18_-_63825408 | 4.79 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr8_-_59226597 | 4.76 |
ENSRNOT00000088208
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_8989580 | 4.76 |
ENSRNOT00000061402
|
Thrb
|
thyroid hormone receptor beta |
| chr6_+_94835845 | 4.71 |
ENSRNOT00000006321
|
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr5_-_77749613 | 4.70 |
ENSRNOT00000075988
|
Mup5
|
major urinary protein 5 |
| chr9_+_44491177 | 4.66 |
ENSRNOT00000075242
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr1_+_78417719 | 4.63 |
ENSRNOT00000020609
|
Tmem160
|
transmembrane protein 160 |
| chr3_-_103460529 | 4.63 |
ENSRNOT00000047274
|
LOC100909573
|
olfactory receptor 4F6-like |
| chr9_-_40008680 | 4.59 |
ENSRNOT00000016578
|
Khdrbs2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr9_+_73529612 | 4.58 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_-_143392532 | 4.58 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr9_-_27761365 | 4.57 |
ENSRNOT00000018552
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr5_+_160306727 | 4.54 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr13_-_50549981 | 4.54 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr10_-_109909646 | 4.53 |
ENSRNOT00000074362
ENSRNOT00000088907 |
Dcxr
|
dicarbonyl and L-xylulose reductase |
| chr9_+_47281961 | 4.51 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr3_+_56355431 | 4.50 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr5_-_166116516 | 4.49 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr1_-_112436794 | 4.49 |
ENSRNOT00000019971
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chrX_-_31780425 | 4.49 |
ENSRNOT00000004693
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr6_+_48452369 | 4.49 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr9_+_117583610 | 4.47 |
ENSRNOT00000088647
ENSRNOT00000049426 |
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chrX_-_110230610 | 4.42 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr12_+_18678594 | 4.36 |
ENSRNOT00000087229
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr18_-_15089988 | 4.35 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr3_-_176816114 | 4.34 |
ENSRNOT00000079262
ENSRNOT00000018697 |
Stmn3
|
stathmin 3 |
| chr18_-_3676188 | 4.34 |
ENSRNOT00000073811
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr1_+_212181374 | 4.31 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr10_+_109851854 | 4.31 |
ENSRNOT00000087005
|
Aspscr1
|
ASPSCR1, UBX domain containing tether for SLC2A4 |
| chr5_+_79179417 | 4.30 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr18_-_62476700 | 4.26 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr10_+_65733991 | 4.21 |
ENSRNOT00000013698
|
Slc46a1
|
solute carrier family 46 member 1 |
| chr15_+_42659371 | 4.19 |
ENSRNOT00000091612
|
Clu
|
clusterin |
| chr13_-_82006005 | 4.17 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr19_-_55183557 | 4.16 |
ENSRNOT00000017317
|
Mlnr
|
motilin receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 5.2 | 26.2 | GO:0015755 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 3.9 | 46.8 | GO:0019627 | urea metabolic process(GO:0019627) |
| 3.4 | 13.6 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 3.2 | 12.9 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 3.1 | 21.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 2.9 | 14.5 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 2.8 | 11.2 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 2.6 | 7.8 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 2.5 | 7.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.4 | 7.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 2.4 | 12.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 2.4 | 7.1 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 2.2 | 6.7 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 2.2 | 6.5 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 2.2 | 10.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 2.1 | 6.2 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 2.0 | 9.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.9 | 9.5 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 1.9 | 30.4 | GO:0051923 | sulfation(GO:0051923) |
| 1.8 | 5.4 | GO:0021678 | third ventricle development(GO:0021678) |
| 1.8 | 38.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.7 | 18.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.6 | 6.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.6 | 4.9 | GO:0061235 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 1.6 | 8.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.5 | 7.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 1.5 | 4.5 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.5 | 4.4 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 1.4 | 4.2 | GO:1902988 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 1.4 | 4.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.4 | 19.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.3 | 10.7 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.3 | 11.8 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.2 | 15.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.2 | 5.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 1.1 | 13.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 1.1 | 3.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.1 | 7.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.1 | 4.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 1.1 | 5.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 4.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 1.0 | 11.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.0 | 8.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 1.0 | 4.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.0 | 9.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.0 | 14.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.0 | 4.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.0 | 7.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.0 | 9.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.0 | 2.9 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 1.0 | 3.8 | GO:0042853 | L-alanine catabolic process(GO:0042853) |
| 1.0 | 4.8 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.9 | 3.8 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.9 | 3.8 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.9 | 5.5 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.9 | 4.5 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.9 | 2.7 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.9 | 4.5 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.9 | 10.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.9 | 8.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.9 | 9.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.9 | 6.0 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.8 | 2.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.8 | 4.8 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.8 | 6.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.8 | 6.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.8 | 5.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.7 | 2.2 | GO:0045751 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of Toll signaling pathway(GO:0008592) negative regulation of Toll signaling pathway(GO:0045751) |
| 0.7 | 5.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.7 | 2.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 14.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.7 | 2.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.7 | 4.9 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.7 | 7.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.7 | 2.1 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.7 | 7.6 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.7 | 16.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.7 | 12.5 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.7 | 11.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 7.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.6 | 1.8 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.6 | 8.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 2.3 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.6 | 2.9 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.6 | 22.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.6 | 8.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 1.6 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.5 | 4.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.5 | 4.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.5 | 2.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.5 | 5.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 6.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.5 | 6.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 1.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.5 | 4.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.5 | 3.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.5 | 4.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.5 | 5.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 11.8 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.5 | 1.4 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 1.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.4 | 0.9 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 1.3 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 2.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 2.1 | GO:0002786 | positive regulation of antimicrobial peptide production(GO:0002225) regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 2.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.4 | 1.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 0.4 | 1.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.4 | 2.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.4 | 2.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.4 | 3.5 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 3.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.4 | 1.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 6.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.4 | 2.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.4 | 7.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.4 | 2.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.4 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.4 | 8.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 2.6 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.4 | 1.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 3.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 4.9 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 2.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.3 | 4.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.3 | 2.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 7.5 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.3 | 1.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 12.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.3 | 1.0 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.3 | 1.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.3 | 10.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 1.5 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 2.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 1.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 3.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 1.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 7.8 | GO:0006956 | complement activation(GO:0006956) |
| 0.3 | 2.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 0.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.3 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.7 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.2 | 2.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 1.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.9 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 2.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 3.7 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.2 | 11.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.9 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.2 | 1.8 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.2 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 3.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 1.0 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 3.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 0.8 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 1.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 1.7 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.2 | 1.3 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.2 | 2.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 2.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.2 | 2.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 3.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.9 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 2.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 4.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 1.6 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 0.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.2 | 1.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 3.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 0.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 2.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 2.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 12.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 19.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 5.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 3.4 | GO:1905039 | carboxylic acid transmembrane transport(GO:1905039) |
| 0.1 | 1.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 1.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 3.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 3.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 3.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 3.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 1.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 4.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 163.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 6.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 3.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) |
| 0.1 | 5.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 4.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.6 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.1 | 5.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 1.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.2 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 1.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.1 | 1.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 2.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 8.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 1.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 3.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 18.1 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 5.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 7.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 6.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 4.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.5 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.1 | 1.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.1 | 4.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 1.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 3.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.2 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 1.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.7 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 3.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.8 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.1 | 1.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.9 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 1.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 1.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.9 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 2.2 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.9 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 1.2 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 4.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 2.9 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 7.2 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.1 | 0.3 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 3.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.8 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.2 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.4 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 3.2 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.9 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.4 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 1.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 2.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 4.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 2.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 2.0 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.8 | GO:0072528 | pyrimidine-containing compound biosynthetic process(GO:0072528) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.8 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.3 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 1.2 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 1.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 1.4 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 25.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.5 | 14.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.3 | 6.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.2 | 9.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.0 | 2.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.8 | 14.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.7 | 3.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 7.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 7.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.6 | 4.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 3.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 5.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.6 | 7.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 5.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 2.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 1.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.5 | 6.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.5 | 3.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.5 | 4.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.5 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 4.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 2.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 1.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 12.0 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 11.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 6.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 1.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.4 | 10.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 1.3 | GO:0005962 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.4 | 8.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 7.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.4 | 46.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 1.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.4 | 34.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 2.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 2.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 4.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.4 | 6.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 17.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 10.9 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 1.8 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 7.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 4.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 8.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 10.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 3.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 30.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.3 | 2.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 8.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 2.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 3.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 0.8 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 7.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 3.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 6.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 4.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 10.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 7.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 11.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 0.9 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 3.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 2.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 8.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 1.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 11.9 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 10.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.9 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 4.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 4.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 9.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 2.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.2 | GO:0098862 | cluster of actin-based cell projections(GO:0098862) |
| 0.1 | 39.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 3.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 5.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 12.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 16.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 17.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.6 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 13.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 2.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 16.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 1.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 10.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.6 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 4.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 10.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 6.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 2.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 27.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 4.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 4.1 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.8 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 17.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 76.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 4.5 | 13.6 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 3.8 | 11.3 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 3.6 | 14.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 3.4 | 23.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.3 | 9.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.9 | 26.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 2.7 | 73.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 2.7 | 8.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 2.5 | 17.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.1 | 10.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.8 | 9.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.8 | 7.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.6 | 6.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.6 | 4.9 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.6 | 7.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.6 | 48.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.5 | 18.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.5 | 9.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 1.5 | 7.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.4 | 21.7 | GO:0005542 | folic acid binding(GO:0005542) |
| 1.4 | 7.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.4 | 4.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.4 | 5.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.3 | 6.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.3 | 3.8 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.3 | 2.6 | GO:0070905 | serine binding(GO:0070905) |
| 1.3 | 6.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.2 | 6.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.2 | 7.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.2 | 3.7 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 1.2 | 9.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.2 | 20.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.2 | 3.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 1.1 | 8.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.1 | 14.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.1 | 18.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 1.1 | 12.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.0 | 3.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.0 | 14.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.9 | 7.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.9 | 2.7 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.8 | 4.8 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.8 | 4.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 7.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.8 | 5.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.7 | 4.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.7 | 6.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 7.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.7 | 8.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 7.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.7 | 3.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.6 | 6.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.6 | 11.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.6 | 16.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.6 | 7.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.6 | 12.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.6 | 3.5 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.6 | 3.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.6 | 2.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 6.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.5 | 4.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 1.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 11.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.5 | 3.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.5 | 4.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 1.5 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.5 | 10.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.5 | 2.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.5 | 2.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.5 | 2.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 2.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 8.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 5.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 2.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.4 | 2.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 6.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.4 | 20.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.4 | 3.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 1.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.4 | 5.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.4 | 6.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 3.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.4 | 2.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 4.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.0 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.3 | 1.4 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 9.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 3.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.3 | 2.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 17.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.3 | 3.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 0.9 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 1.7 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 0.8 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 1.9 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 7.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 1.3 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 2.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 2.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 6.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 4.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 5.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 1.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 8.9 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.2 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 3.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 2.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.2 | 1.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.8 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.2 | 21.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.9 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.7 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 2.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 3.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 1.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 3.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 6.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 11.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 3.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 10.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 11.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 163.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 12.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 3.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.7 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.9 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 6.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 14.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 5.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 9.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 8.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.5 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 5.7 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 4.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 6.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 6.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 3.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 1.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 4.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 6.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 4.9 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.8 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 2.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 31.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.6 | 11.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 3.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 12.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 7.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 7.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 1.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 6.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 35.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.6 | 19.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.6 | 26.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.2 | 7.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 1.2 | 19.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 7.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.9 | 8.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 14.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.8 | 10.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 25.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 9.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 7.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 5.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.5 | 6.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 19.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 3.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 24.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.4 | 10.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 22.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 3.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 12.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 3.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 6.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 4.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 17.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 4.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 7.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 4.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 2.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 3.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 7.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 2.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.2 | 4.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 3.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 8.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 8.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 20.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 7.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 7.7 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |