Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
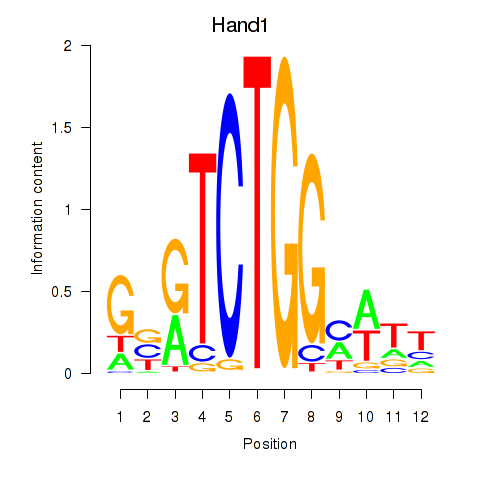
Results for Hand1
Z-value: 0.65
Transcription factors associated with Hand1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hand1
|
ENSRNOG00000002582 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hand1 | rn6_v1_chr10_-_43253296_43253296 | 0.18 | 1.1e-03 | Click! |
Activity profile of Hand1 motif
Sorted Z-values of Hand1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_57952982 | 21.44 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chr1_-_224389389 | 18.06 |
ENSRNOT00000077408
ENSRNOT00000050010 |
UST4r
|
integral membrane transport protein UST4r |
| chr1_+_279867034 | 17.78 |
ENSRNOT00000024164
|
Pnliprp1
|
pancreatic lipase-related protein 1 |
| chr1_-_198486157 | 17.66 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr1_+_65576535 | 16.70 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr7_+_2689501 | 13.52 |
ENSRNOT00000041341
|
Apof
|
apolipoprotein F |
| chr1_-_170431073 | 13.02 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr7_-_71226150 | 12.78 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr6_-_127620296 | 12.45 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr17_-_10818835 | 12.37 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr1_-_253000760 | 12.14 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr16_-_7007051 | 11.42 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_-_4945352 | 11.39 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr8_-_21901829 | 11.14 |
ENSRNOT00000027963
|
Angptl6
|
angiopoietin-like 6 |
| chr4_-_161850875 | 10.94 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr16_-_7007287 | 10.93 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chrX_+_71272042 | 10.18 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr5_-_78985990 | 9.96 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr18_-_24057917 | 9.39 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr5_-_34813116 | 9.36 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr1_-_216080287 | 9.31 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr16_-_7026540 | 8.94 |
ENSRNOT00000051751
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr12_-_46889082 | 8.85 |
ENSRNOT00000001525
|
Pla2g1b
|
phospholipase A2 group IB |
| chr16_+_50152008 | 8.80 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr1_-_224533219 | 8.72 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr1_+_83163079 | 8.60 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr5_+_120340646 | 8.29 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr8_-_50526843 | 7.94 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr9_+_73334618 | 7.82 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr6_-_127632265 | 7.79 |
ENSRNOT00000084157
|
Serpina1
|
serpin family A member 1 |
| chr20_+_31892515 | 7.76 |
ENSRNOT00000071394
|
Tacr2
|
tachykinin receptor 2 |
| chr9_-_4168221 | 7.63 |
ENSRNOT00000061861
|
Sult1c2a
|
sulfotransferase family, cytosolic, 1C, member 2a |
| chr4_+_166601960 | 7.57 |
ENSRNOT00000073394
|
AABR07062266.1
|
|
| chr7_+_26375866 | 7.51 |
ENSRNOT00000059639
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr1_+_105285419 | 7.16 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr1_+_219833299 | 7.05 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr1_+_217173199 | 6.98 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_+_31493107 | 6.58 |
ENSRNOT00000023611
ENSRNOT00000086264 |
Tubb2a
|
tubulin, beta 2A class IIa |
| chr1_+_100199057 | 6.49 |
ENSRNOT00000025831
|
Klk1
|
kallikrein 1 |
| chr9_-_32019205 | 6.38 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr6_-_15191660 | 6.19 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr16_+_22361998 | 6.03 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr17_+_12762752 | 6.02 |
ENSRNOT00000044842
|
Diras2
|
DIRAS family GTPase 2 |
| chr1_+_266952561 | 5.96 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr2_+_20857202 | 5.89 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr1_+_83653234 | 5.76 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr1_-_101449829 | 5.73 |
ENSRNOT00000028315
|
LOC100360087
|
ferritin light chain 1-like |
| chrX_+_1311121 | 5.59 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr3_-_176816114 | 5.53 |
ENSRNOT00000079262
ENSRNOT00000018697 |
Stmn3
|
stathmin 3 |
| chr19_+_15143794 | 5.52 |
ENSRNOT00000085762
|
Ces1f
|
carboxylesterase 1F |
| chr5_-_78376032 | 5.47 |
ENSRNOT00000075916
|
Alad
|
aminolevulinate dehydratase |
| chr12_-_41448668 | 5.31 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr18_+_63203063 | 5.27 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr16_+_20432899 | 5.27 |
ENSRNOT00000026271
|
Mpv17l2
|
MPV17 mitochondrial inner membrane protein like 2 |
| chr11_+_9642365 | 5.24 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr1_-_125967756 | 5.20 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_+_95221474 | 5.19 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr7_+_93975451 | 5.14 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chrX_-_54303729 | 5.14 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr1_+_201620642 | 5.09 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_+_218569510 | 5.08 |
ENSRNOT00000019652
|
Cpt1a
|
carnitine palmitoyltransferase 1A |
| chr11_+_77815181 | 5.05 |
ENSRNOT00000002640
|
Cldn1
|
claudin 1 |
| chr4_-_117575154 | 5.02 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr15_+_28028521 | 4.99 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr10_-_70788309 | 4.94 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr1_+_256955652 | 4.90 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr1_+_198383201 | 4.87 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr1_-_212549477 | 4.86 |
ENSRNOT00000024765
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr1_+_202432366 | 4.65 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr4_-_117568348 | 4.61 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr11_+_66316606 | 4.37 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr20_-_28263037 | 4.36 |
ENSRNOT00000030348
|
Edar
|
ectodysplasin-A receptor |
| chr3_-_172537877 | 4.34 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr1_-_22269831 | 4.33 |
ENSRNOT00000081804
|
Stx7
|
syntaxin 7 |
| chrX_-_110230610 | 4.32 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chrX_+_151103576 | 4.18 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr10_+_74874130 | 4.13 |
ENSRNOT00000076050
ENSRNOT00000044226 ENSRNOT00000085052 ENSRNOT00000076196 |
Sept4
|
septin 4 |
| chr8_+_29453643 | 4.12 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr11_-_77593171 | 4.12 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr2_+_54466280 | 4.05 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr7_-_1738642 | 4.03 |
ENSRNOT00000075554
|
AABR07055356.1
|
|
| chr4_+_86275717 | 4.01 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr1_+_145715969 | 3.88 |
ENSRNOT00000037996
|
Tmc3
|
transmembrane channel-like 3 |
| chr2_-_102370757 | 3.85 |
ENSRNOT00000074255
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr3_+_121490812 | 3.82 |
ENSRNOT00000023913
|
AABR07053707.1
|
|
| chr13_-_91872954 | 3.75 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr7_+_3216497 | 3.68 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr2_+_217963456 | 3.61 |
ENSRNOT00000024243
|
Olfm3
|
olfactomedin 3 |
| chr14_+_2613406 | 3.56 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr1_+_217345154 | 3.56 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_39266959 | 3.56 |
ENSRNOT00000046590
|
Ei24
|
EI24, autophagy associated transmembrane protein |
| chr1_-_275906686 | 3.55 |
ENSRNOT00000092170
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr5_+_139790395 | 3.54 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr16_+_74886719 | 3.51 |
ENSRNOT00000089265
|
Atp7b
|
ATPase copper transporting beta |
| chr17_-_43584152 | 3.48 |
ENSRNOT00000023241
|
Slc17a2
|
solute carrier family 17, member 2 |
| chr7_-_29152442 | 3.48 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr1_-_188407132 | 3.46 |
ENSRNOT00000073233
|
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr1_+_100131449 | 3.44 |
ENSRNOT00000091964
ENSRNOT00000071416 |
Klk1
|
kallikrein 1 |
| chr5_+_120568242 | 3.40 |
ENSRNOT00000050597
|
Lepr
|
leptin receptor |
| chr10_-_52710862 | 3.40 |
ENSRNOT00000005583
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr10_-_38838272 | 3.38 |
ENSRNOT00000089495
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr3_+_121235119 | 3.38 |
ENSRNOT00000023419
|
Mertk
|
MER proto-oncogene, tyrosine kinase |
| chr19_-_601469 | 3.31 |
ENSRNOT00000016462
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr16_+_17493953 | 3.30 |
ENSRNOT00000014819
|
Fam213a
|
family with sequence similarity 213, member A |
| chr1_+_201672528 | 3.27 |
ENSRNOT00000093490
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr11_+_61662270 | 3.24 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr2_-_188138177 | 3.23 |
ENSRNOT00000027474
|
Syt11
|
synaptotagmin 11 |
| chr19_-_21492468 | 3.20 |
ENSRNOT00000087660
ENSRNOT00000048345 ENSRNOT00000020770 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr4_-_31730386 | 3.14 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr10_+_35726544 | 3.14 |
ENSRNOT00000004323
|
Mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr17_+_43661222 | 3.13 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr1_+_224824799 | 3.12 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr5_-_147828449 | 3.11 |
ENSRNOT00000083369
|
Ccdc28b
|
coiled coil domain containing 28B |
| chr18_-_29629087 | 3.02 |
ENSRNOT00000039291
|
Hars
|
histidyl-tRNA synthetase |
| chr3_-_2853272 | 2.97 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr11_-_70499200 | 2.96 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr7_+_2300434 | 2.91 |
ENSRNOT00000071785
|
AABR07055413.1
|
|
| chr3_-_8766433 | 2.88 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr1_-_89358166 | 2.82 |
ENSRNOT00000044532
|
Mag
|
myelin-associated glycoprotein |
| chr3_-_3541947 | 2.82 |
ENSRNOT00000024786
|
Nacc2
|
NACC family member 2 |
| chr1_-_88826302 | 2.78 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr3_-_57957346 | 2.78 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr1_-_23556241 | 2.76 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr15_-_28044210 | 2.69 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chrX_+_80213332 | 2.69 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr8_+_130538651 | 2.69 |
ENSRNOT00000026343
|
Ackr2
|
atypical chemokine receptor 2 |
| chr1_-_53520788 | 2.68 |
ENSRNOT00000060121
|
Gpr31
|
G protein-coupled receptor 31 |
| chr16_+_7303578 | 2.67 |
ENSRNOT00000025656
|
Sema3g
|
semaphorin 3G |
| chr1_-_80783898 | 2.66 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr4_-_117584819 | 2.65 |
ENSRNOT00000090815
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr1_+_154131926 | 2.64 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr1_-_80056574 | 2.63 |
ENSRNOT00000021200
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr2_-_210874304 | 2.62 |
ENSRNOT00000088657
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr5_-_169160987 | 2.62 |
ENSRNOT00000055487
|
Thap3
|
THAP domain containing 3 |
| chr17_+_10384511 | 2.61 |
ENSRNOT00000024357
|
Sncb
|
synuclein, beta |
| chrX_-_136303249 | 2.58 |
ENSRNOT00000093185
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr9_+_100380515 | 2.55 |
ENSRNOT00000022290
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr14_-_33677031 | 2.54 |
ENSRNOT00000002908
|
RGD1311575
|
hypothetical LOC289568 |
| chr2_-_188561267 | 2.50 |
ENSRNOT00000089781
ENSRNOT00000092093 |
Trim46
|
tripartite motif-containing 46 |
| chr10_-_11760620 | 2.50 |
ENSRNOT00000009283
|
Dnase1
|
deoxyribonuclease 1 |
| chr8_-_112884062 | 2.47 |
ENSRNOT00000016222
ENSRNOT00000085585 |
Acpp
|
acid phosphatase, prostate |
| chr2_-_244370983 | 2.47 |
ENSRNOT00000021458
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr5_-_85123829 | 2.46 |
ENSRNOT00000007578
|
Brinp1
|
BMP/retinoic acid inducible neural specific 1 |
| chr7_-_58149061 | 2.44 |
ENSRNOT00000005157
|
Tph2
|
tryptophan hydroxylase 2 |
| chr11_-_87762301 | 2.43 |
ENSRNOT00000000312
|
Car15
|
carbonic anhydrase 15 |
| chr1_-_112436794 | 2.41 |
ENSRNOT00000019971
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr3_-_51297852 | 2.41 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_149333597 | 2.40 |
ENSRNOT00000066948
|
P2ry14
|
purinergic receptor P2Y14 |
| chr15_-_48445588 | 2.40 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr2_+_185393441 | 2.36 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr2_+_198303168 | 2.34 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr13_-_51992693 | 2.32 |
ENSRNOT00000008282
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr9_-_85617954 | 2.28 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr14_-_2032593 | 2.24 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr6_-_98695988 | 2.23 |
ENSRNOT00000006913
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr15_-_9086282 | 2.22 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr4_-_59809321 | 2.20 |
ENSRNOT00000017536
|
Plxna4
|
plexin A4 |
| chr3_-_11410732 | 2.20 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr18_-_58423196 | 2.17 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr11_-_14160697 | 2.17 |
ENSRNOT00000081096
|
Hspa13
|
heat shock protein 70 family, member 13 |
| chr4_-_156427755 | 2.15 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr10_-_40805941 | 2.14 |
ENSRNOT00000017588
|
Atox1
|
antioxidant 1 copper chaperone |
| chr13_-_110678389 | 2.14 |
ENSRNOT00000082906
|
AABR07022168.1
|
|
| chr4_-_157008947 | 2.12 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr4_-_117767772 | 2.10 |
ENSRNOT00000084170
|
LOC103690120
|
probable N-acetyltransferase CML1 |
| chr1_+_65522118 | 2.09 |
ENSRNOT00000058563
|
Mzf1
|
myeloid zinc finger 1 |
| chrX_+_62727755 | 2.09 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr5_+_157434481 | 2.06 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr1_+_167937026 | 2.01 |
ENSRNOT00000020655
|
Olr57
|
olfactory receptor 57 |
| chr13_+_99173484 | 1.99 |
ENSRNOT00000080574
ENSRNOT00000088654 |
Lefty2
|
Left-right determination factor 2 |
| chrX_+_119030419 | 1.99 |
ENSRNOT00000060168
|
Pls3
|
plastin 3 |
| chr6_-_99843245 | 1.95 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr8_+_70603249 | 1.95 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr8_-_76579387 | 1.93 |
ENSRNOT00000090747
|
Fam81a
|
family with sequence similarity 81, member A |
| chr14_+_60657686 | 1.93 |
ENSRNOT00000070892
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr14_-_14390699 | 1.93 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr16_-_37159745 | 1.92 |
ENSRNOT00000084469
|
Fbxo8
|
F-box protein 8 |
| chr10_-_88677055 | 1.92 |
ENSRNOT00000025590
|
Ghdc
|
GH3 domain containing |
| chr8_-_76579099 | 1.89 |
ENSRNOT00000088628
|
Fam81a
|
family with sequence similarity 81, member A |
| chr5_-_12526962 | 1.89 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr20_+_1749716 | 1.87 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr1_+_94579080 | 1.87 |
ENSRNOT00000020079
|
LOC690000
|
similar to CG3740-PA |
| chr7_-_139223116 | 1.86 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr1_-_220188541 | 1.86 |
ENSRNOT00000026968
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr6_-_108415093 | 1.85 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr6_-_142880382 | 1.82 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr5_-_136721379 | 1.82 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr16_+_83288664 | 1.82 |
ENSRNOT00000019993
|
Cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr9_+_86874685 | 1.81 |
ENSRNOT00000041337
|
NEWGENE_1305560
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr2_+_202470487 | 1.81 |
ENSRNOT00000026953
|
Gdap2
|
ganglioside-induced differentiation-associated-protein 2 |
| chr2_+_144646308 | 1.79 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr5_+_144281614 | 1.78 |
ENSRNOT00000014383
|
Trappc3
|
trafficking protein particle complex 3 |
| chr1_+_44446765 | 1.75 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr10_-_110431792 | 1.74 |
ENSRNOT00000054922
|
LOC619574
|
hypothetical protein LOC619574 |
| chr1_+_78417719 | 1.73 |
ENSRNOT00000020609
|
Tmem160
|
transmembrane protein 160 |
| chr11_+_28692708 | 1.72 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr6_+_86823684 | 1.72 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr20_-_2210033 | 1.71 |
ENSRNOT00000084596
|
Trim26
|
tripartite motif-containing 26 |
| chrX_+_106306795 | 1.70 |
ENSRNOT00000073661
|
Gprasp1
|
G protein-coupled receptor associated sorting protein 1 |
| chr2_+_250600823 | 1.68 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hand1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0033986 | response to methanol(GO:0033986) |
| 4.2 | 16.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 3.3 | 13.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 2.9 | 8.8 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 2.6 | 15.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 2.5 | 7.5 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.0 | 8.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 2.0 | 21.8 | GO:0015747 | urate transport(GO:0015747) |
| 1.9 | 7.8 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 1.9 | 5.7 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 1.8 | 7.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.7 | 5.2 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 1.7 | 5.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.7 | 5.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 1.5 | 5.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.4 | 7.2 | GO:0036233 | glycine import(GO:0036233) |
| 1.4 | 4.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 1.4 | 4.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.3 | 9.4 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 1.3 | 8.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.2 | 6.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.2 | 2.4 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 1.2 | 19.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.1 | 4.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.1 | 3.2 | GO:1903422 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.1 | 5.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 1.1 | 10.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.0 | 3.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.0 | 12.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.9 | 31.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.9 | 6.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.9 | 5.5 | GO:0010266 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 0.9 | 2.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.9 | 3.4 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of locomotor rhythm(GO:1904059) |
| 0.8 | 4.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.8 | 2.5 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.8 | 5.8 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.7 | 3.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.7 | 4.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.7 | 2.2 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.7 | 8.6 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.7 | 3.3 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 0.7 | 2.0 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.6 | 5.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.6 | 2.9 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.6 | 5.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.6 | 3.4 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.6 | 5.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 2.2 | GO:0021615 | vagus nerve development(GO:0021564) glossopharyngeal nerve morphogenesis(GO:0021615) chemorepulsion of axon(GO:0061643) |
| 0.5 | 1.6 | GO:0046154 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.5 | 1.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.5 | 2.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.5 | 16.5 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.5 | 3.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.5 | 7.2 | GO:0015865 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.5 | 1.4 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.5 | 6.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.5 | 1.4 | GO:0046687 | response to chromate(GO:0046687) |
| 0.4 | 2.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 8.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 3.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 0.8 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.4 | 1.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) unidimensional cell growth(GO:0009826) axial mesoderm morphogenesis(GO:0048319) |
| 0.4 | 2.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.4 | 3.7 | GO:0001554 | luteolysis(GO:0001554) |
| 0.4 | 1.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.4 | 3.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 3.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.3 | 13.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.3 | 10.5 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.3 | 1.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 2.5 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.3 | 0.3 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.3 | 1.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 1.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 1.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.3 | 0.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.3 | 1.6 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 1.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.3 | 0.5 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.3 | 2.3 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.3 | 4.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 1.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 1.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 1.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 2.1 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.2 | 2.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 2.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 0.7 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.2 | 1.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 4.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 2.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 7.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 4.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 2.3 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 | 4.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 17.1 | GO:0031016 | pancreas development(GO:0031016) |
| 0.2 | 5.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 2.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 11.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.2 | 2.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 5.9 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.2 | 0.5 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 2.5 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.6 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 0.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.5 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 4.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:0045077 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 3.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 1.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 3.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 1.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.7 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 4.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.3 | GO:2000525 | type IV hypersensitivity(GO:0001806) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 3.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.9 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 1.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 2.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 2.7 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 1.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.5 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 1.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 2.6 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 4.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 2.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.3 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 5.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.4 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 2.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.9 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 1.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 4.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 2.8 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 4.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 2.2 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 3.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.8 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 1.5 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 7.0 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 4.5 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 3.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.6 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 3.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.2 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 2.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.9 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.7 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 2.1 | 12.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.1 | 13.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.0 | 9.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.9 | 7.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.7 | 10.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.7 | 4.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 5.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.6 | 4.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 4.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 8.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 2.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 1.5 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.5 | 1.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 1.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 3.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 9.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 4.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 6.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 6.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 16.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 3.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 5.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 30.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.2 | 1.7 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 2.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 3.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 5.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 4.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 1.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.6 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 6.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 3.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 4.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.1 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 11.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 14.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 8.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 4.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 55.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 37.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 5.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 22.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 6.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.4 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 2.6 | 7.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 2.4 | 7.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 2.3 | 7.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 2.2 | 10.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 2.1 | 20.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 2.0 | 6.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 2.0 | 10.0 | GO:0019862 | IgA binding(GO:0019862) |
| 1.8 | 5.5 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 1.8 | 18.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.8 | 5.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.7 | 5.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.7 | 11.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.5 | 5.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.4 | 13.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.4 | 4.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.3 | 5.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.2 | 8.4 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.2 | 5.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 3.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 1.1 | 2.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 1.0 | 8.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.0 | 12.8 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 1.0 | 2.9 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.9 | 7.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.9 | 2.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.9 | 17.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.9 | 6.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 12.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.9 | 4.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.8 | 4.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.8 | 21.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 2.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.7 | 10.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.7 | 9.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 3.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.7 | 3.5 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.7 | 2.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 4.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.5 | 4.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 14.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 5.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 50.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 3.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.4 | 2.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 12.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 3.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 2.6 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.4 | 4.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 7.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 4.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 1.4 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.3 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 2.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 4.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.3 | 1.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 2.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 3.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 1.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 5.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 2.5 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 3.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 2.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 2.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 3.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 2.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.2 | 2.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 5.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 2.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.6 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 1.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 7.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 4.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 6.2 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 13.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 2.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 4.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 3.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.5 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.1 | 1.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 7.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.4 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 3.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 13.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 1.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 1.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 5.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 20.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 9.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 9.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 6.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 6.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 45.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 5.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 7.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 5.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 5.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 10.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 15.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 16.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.7 | 11.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 13.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 8.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.5 | 9.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.5 | 12.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.5 | 9.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.4 | 8.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 6.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 13.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 5.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 5.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 4.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 5.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 2.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 6.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 18.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 0.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 7.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 2.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 4.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.5 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 3.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 3.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.4 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 3.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |