Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.62
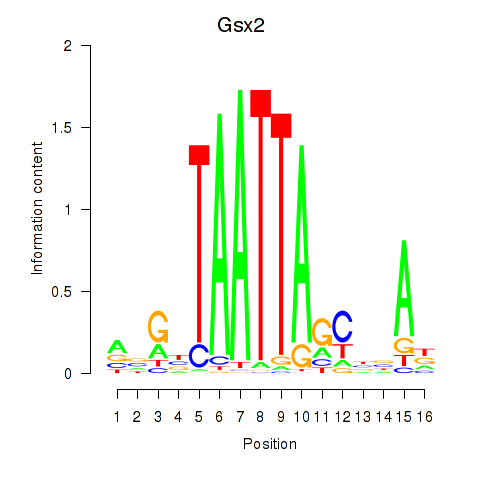
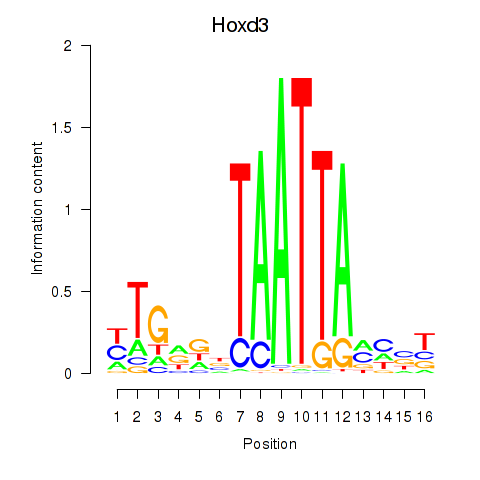
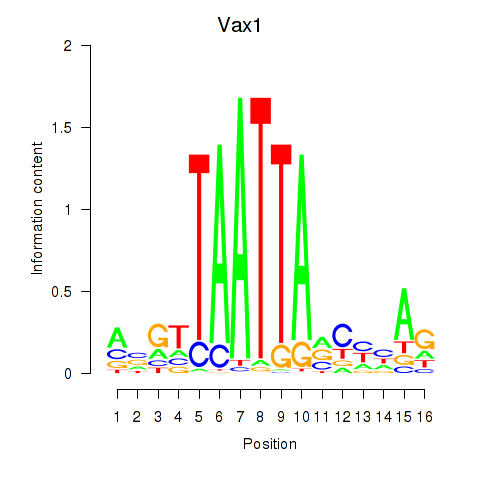
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSRNOG00000002266 | GS homeobox 2 |
|
Hoxd3
|
ENSRNOG00000001577 | homeo box D3 |
|
Vax1
|
ENSRNOG00000008824 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd3 | rn6_v1_chr3_+_61658245_61658245 | 0.32 | 7.6e-09 | Click! |
| Vax1 | rn6_v1_chr1_-_280338813_280338813 | 0.07 | 1.9e-01 | Click! |
| Gsx2 | rn6_v1_chr14_-_35652709_35652709 | -0.02 | 7.8e-01 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_76488216 | 29.77 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr8_+_33239139 | 27.89 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_-_158133861 | 26.36 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_87224677 | 24.02 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr17_-_87826421 | 18.94 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr5_-_161981441 | 18.02 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr8_-_7426611 | 12.02 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr2_-_138833933 | 11.80 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr2_-_57935334 | 11.52 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr9_+_73418607 | 11.13 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr12_-_19167015 | 11.13 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr18_+_30036887 | 9.96 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr7_-_101138860 | 9.52 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr17_+_11683862 | 8.75 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr5_-_168734296 | 8.60 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_+_59156251 | 8.59 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr2_-_158156444 | 8.37 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_266315036 | 7.89 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr7_-_101138373 | 7.83 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr6_+_2216623 | 7.63 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrX_+_6273733 | 7.61 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr18_+_32336102 | 7.43 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr5_-_17061837 | 7.34 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr2_-_40386669 | 7.02 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr11_-_62067655 | 6.93 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_217345545 | 6.73 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chrX_+_14019961 | 6.50 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_104291071 | 6.48 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr6_+_8284878 | 6.37 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr2_+_252090669 | 6.34 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr18_-_26656879 | 6.20 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_-_154627257 | 6.12 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr2_-_238369692 | 5.93 |
ENSRNOT00000041583
|
Arhgef38
|
Rho guanine nucleotide exchange factor 38 |
| chr7_-_143497108 | 5.91 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chrX_+_84064427 | 5.87 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr9_-_44419998 | 5.67 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr11_+_36851038 | 5.65 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr2_+_145174876 | 5.63 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr5_-_17061361 | 5.58 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr17_-_84247038 | 5.52 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr2_+_158097843 | 5.49 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr2_+_54466280 | 5.31 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr2_+_66940057 | 5.31 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr5_+_64476317 | 5.27 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr9_-_44237117 | 5.23 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr2_-_219262901 | 5.16 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr18_+_51523758 | 5.06 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr8_-_78233430 | 5.01 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrX_+_65566047 | 4.95 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr17_-_14627937 | 4.89 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr2_-_60657712 | 4.87 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr6_-_144123596 | 4.65 |
ENSRNOT00000006144
ENSRNOT00000087845 |
Wdr60
|
WD repeat domain 60 |
| chr6_+_104291340 | 4.64 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr16_-_10802512 | 4.64 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr18_-_24735349 | 4.55 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr3_+_15560712 | 4.54 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr20_+_13817795 | 4.50 |
ENSRNOT00000036518
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr1_-_256734719 | 4.47 |
ENSRNOT00000021546
ENSRNOT00000089456 |
Myof
|
myoferlin |
| chr8_+_44136496 | 4.35 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr4_+_139670092 | 4.31 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr4_-_84740909 | 4.27 |
ENSRNOT00000013088
|
Scrn1
|
secernin 1 |
| chr4_-_72143748 | 4.20 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr10_+_54156649 | 3.96 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr1_-_236900904 | 3.95 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr2_+_202200797 | 3.91 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr4_+_158088505 | 3.88 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr20_-_30947484 | 3.81 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr10_-_51778939 | 3.80 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr12_-_6078411 | 3.80 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr4_-_100252755 | 3.70 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr5_-_147412705 | 3.53 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr18_+_30435119 | 3.53 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr5_+_18901039 | 3.53 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr14_-_82975263 | 3.49 |
ENSRNOT00000024165
|
Slc5a1
|
solute carrier family 5 member 1 |
| chrX_-_69218526 | 3.43 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr1_-_224698514 | 3.42 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr4_-_62860446 | 3.41 |
ENSRNOT00000015752
|
Fam180a
|
family with sequence similarity 180, member A |
| chr16_-_3765917 | 3.38 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr3_-_46942966 | 3.28 |
ENSRNOT00000087439
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_143656793 | 3.09 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr1_-_67065797 | 3.03 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr10_-_82117109 | 3.02 |
ENSRNOT00000079711
|
Abcc3
|
ATP binding cassette subfamily C member 3 |
| chr3_+_5709236 | 3.00 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr9_-_85243001 | 2.97 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr14_-_45859908 | 2.96 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr3_+_148654668 | 2.95 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr14_+_39964588 | 2.91 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr7_+_26256459 | 2.88 |
ENSRNOT00000010986
|
Appl2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr1_-_215033460 | 2.81 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr9_+_67234303 | 2.79 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr2_-_187113717 | 2.75 |
ENSRNOT00000020147
|
Lrrc71
|
leucine rich repeat containing 71 |
| chr2_+_248398917 | 2.71 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr11_+_70056624 | 2.71 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr8_-_102149912 | 2.66 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr8_-_33121002 | 2.63 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr1_+_48077033 | 2.62 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr1_+_168575090 | 2.57 |
ENSRNOT00000048299
|
Olr103
|
olfactory receptor 103 |
| chr11_-_61234944 | 2.52 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr1_-_101095594 | 2.49 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr1_+_99505677 | 2.41 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr6_+_95205153 | 2.41 |
ENSRNOT00000007339
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr1_-_23556241 | 2.40 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr12_+_2140203 | 2.37 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr2_-_33025271 | 2.34 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_84835060 | 2.32 |
ENSRNOT00000007867
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr5_-_133959447 | 2.32 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr7_-_107203897 | 2.30 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr3_+_159368273 | 2.30 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_52664209 | 2.26 |
ENSRNOT00000065126
ENSRNOT00000079020 |
Scn9a
|
sodium voltage-gated channel alpha subunit 9 |
| chr10_+_103395511 | 2.21 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr15_+_11298478 | 2.18 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr6_-_3355339 | 2.14 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_+_80279706 | 2.13 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr4_-_176909075 | 2.12 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr8_-_17525906 | 2.09 |
ENSRNOT00000007855
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr7_+_6644643 | 2.07 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr4_+_87608301 | 1.98 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr13_+_60619309 | 1.97 |
ENSRNOT00000082129
|
AABR07021204.1
|
|
| chr13_+_82355886 | 1.97 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr2_-_96509424 | 1.95 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr1_-_72184265 | 1.90 |
ENSRNOT00000047856
|
Vom1r36
|
vomeronasal 1 receptor 36 |
| chr14_-_77810147 | 1.88 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr10_+_104582955 | 1.87 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr15_+_108526014 | 1.87 |
ENSRNOT00000017211
|
Tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr13_+_82355471 | 1.85 |
ENSRNOT00000030677
|
Sele
|
selectin E |
| chr2_-_181900856 | 1.84 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr18_+_17043903 | 1.84 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr7_+_72924799 | 1.83 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr14_+_34446616 | 1.82 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr4_-_55011415 | 1.67 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr3_-_14643897 | 1.63 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr1_-_275882444 | 1.63 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr20_+_3230052 | 1.57 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr2_-_25235275 | 1.56 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr12_+_17253791 | 1.54 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr12_-_52658275 | 1.54 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr6_+_29977797 | 1.51 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr15_+_45712821 | 1.49 |
ENSRNOT00000083381
ENSRNOT00000045833 |
Fam124a
|
family with sequence similarity 124 member A |
| chr8_+_59344083 | 1.46 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chr7_+_71157664 | 1.41 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_-_5101177 | 1.38 |
ENSRNOT00000002888
|
Lrrc8d
|
leucine rich repeat containing 8 family, member D |
| chr3_-_3594475 | 1.35 |
ENSRNOT00000064861
|
RGD1564379
|
RGD1564379 |
| chr7_-_14364178 | 1.34 |
ENSRNOT00000090673
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr2_-_93985378 | 1.33 |
ENSRNOT00000075493
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr20_+_3246739 | 1.33 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr3_-_111087347 | 1.33 |
ENSRNOT00000018277
|
Rhov
|
ras homolog family member V |
| chrX_-_152642531 | 1.29 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr10_+_86399827 | 1.29 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr9_+_84203072 | 1.20 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr3_+_117421604 | 1.18 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr10_+_64174931 | 1.17 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr5_+_129756149 | 1.13 |
ENSRNOT00000067277
|
Dmrta2
|
DMRT-like family A2 |
| chr1_+_86429262 | 1.12 |
ENSRNOT00000045789
|
Vom1r3
|
vomeronasal 1 receptor 3 |
| chr2_+_243577082 | 1.12 |
ENSRNOT00000016556
|
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr12_+_19328957 | 1.10 |
ENSRNOT00000033288
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr1_+_75298364 | 1.05 |
ENSRNOT00000018981
|
Vom1r61
|
vomeronasal 1 receptor 61 |
| chr1_+_170205591 | 1.05 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr4_-_88684415 | 1.03 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr8_+_41302631 | 1.02 |
ENSRNOT00000076963
|
LOC100910822
|
olfactory receptor 143-like |
| chr3_-_160561741 | 1.00 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr3_-_76102782 | 0.98 |
ENSRNOT00000007690
|
Olr602
|
olfactory receptor 602 |
| chr10_-_47546037 | 0.96 |
ENSRNOT00000066109
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr1_+_277355619 | 0.96 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr15_-_54528480 | 0.93 |
ENSRNOT00000066888
|
Fndc3a
|
fibronectin type III domain containing 3a |
| chrX_+_68627313 | 0.89 |
ENSRNOT00000076699
ENSRNOT00000076795 ENSRNOT00000008705 |
Yipf6
|
Yip1 domain family, member 6 |
| chr6_-_104290579 | 0.88 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr5_+_58995249 | 0.88 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr14_-_82658891 | 0.87 |
ENSRNOT00000006841
|
Uvssa
|
UV-stimulated scaffold protein A |
| chr4_+_6827429 | 0.87 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr13_-_89545182 | 0.86 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chrM_+_9870 | 0.83 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr17_-_54678710 | 0.83 |
ENSRNOT00000046013
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr1_-_190370499 | 0.80 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr6_-_127319362 | 0.80 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr7_-_55604403 | 0.79 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr17_+_45801528 | 0.78 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr15_-_93765498 | 0.76 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr10_-_47546345 | 0.72 |
ENSRNOT00000077461
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr10_-_52290657 | 0.71 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr7_-_3229167 | 0.69 |
ENSRNOT00000008929
|
Tmem198b
|
transmembrane protein 198b |
| chr14_-_24123253 | 0.67 |
ENSRNOT00000002743
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr4_-_51199570 | 0.64 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr2_+_85305225 | 0.64 |
ENSRNOT00000015904
|
Tas2r119
|
taste receptor, type 2, member 119 |
| chr3_-_114251647 | 0.63 |
ENSRNOT00000024245
|
Duoxa1
|
dual oxidase maturation factor 1 |
| chrX_+_37329779 | 0.61 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr5_-_28164326 | 0.60 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr8_+_18795525 | 0.60 |
ENSRNOT00000050430
|
Olr1124
|
olfactory receptor 1124 |
| chr3_+_102947730 | 0.58 |
ENSRNOT00000071260
|
Olr773
|
olfactory receptor 773 |
| chr9_+_10013854 | 0.57 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr14_-_6533524 | 0.56 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr14_+_70164650 | 0.55 |
ENSRNOT00000004385
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr2_+_187951344 | 0.54 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr9_+_95295701 | 0.54 |
ENSRNOT00000025045
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr12_-_37538403 | 0.53 |
ENSRNOT00000001402
|
Snrnp35
|
small nuclear ribonucleoprotein U11/U12 subunit 35 |
| chr8_+_41336340 | 0.52 |
ENSRNOT00000072049
|
Olr1225
|
olfactory receptor 1225 |
| chr3_+_72134731 | 0.51 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr7_-_15821927 | 0.49 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr5_+_142986526 | 0.49 |
ENSRNOT00000012811
|
Rspo1
|
R-spondin 1 |
| chr18_-_16543992 | 0.46 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr9_+_15513063 | 0.44 |
ENSRNOT00000020499
|
Taf8
|
TATA-box binding protein associated factor 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 4.3 | 12.9 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 4.0 | 12.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 2.9 | 8.8 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 2.9 | 11.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.6 | 7.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.3 | 18.0 | GO:0015884 | folic acid transport(GO:0015884) |
| 2.0 | 6.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.8 | 5.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.8 | 5.3 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.5 | 4.6 | GO:0048372 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 1.5 | 7.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.4 | 5.6 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 1.4 | 7.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.4 | 5.5 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.3 | 7.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.3 | 3.8 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.2 | 3.7 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.2 | 4.6 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) |
| 1.0 | 5.2 | GO:0061743 | motor learning(GO:0061743) |
| 1.0 | 3.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.0 | 3.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 4.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 3.5 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.7 | 2.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.6 | 4.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.6 | 4.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.6 | 2.3 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 0.6 | 1.7 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.5 | 1.6 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.5 | 2.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.5 | 6.7 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.5 | 3.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.5 | 2.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 3.9 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 2.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 5.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.4 | 1.2 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.4 | 1.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.4 | 2.7 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.4 | 11.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.4 | 4.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.4 | 6.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 3.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 2.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 3.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 8.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 11.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 29.8 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.3 | 1.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 3.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 2.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 1.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 4.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 1.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 24.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.2 | 30.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.6 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 3.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 1.3 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 1.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 0.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 2.5 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 3.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 2.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 4.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 2.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 3.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 3.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 2.5 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 1.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 6.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 8.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.6 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 2.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 4.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.3 | GO:1904742 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.9 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.6 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 1.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 11.1 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 1.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 2.7 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.5 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.1 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.5 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.0 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 4.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.1 | 2.3 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 22.9 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 3.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 5.9 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0090042 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 4.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.9 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 2.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 2.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 3.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.2 | GO:0030031 | cell projection assembly(GO:0030031) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 12.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.2 | 11.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.2 | 3.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.2 | 4.7 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.0 | 17.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.8 | 3.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.8 | 12.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 5.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 3.0 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.7 | 2.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.6 | 3.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 11.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 2.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 6.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 8.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 32.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 9.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 12.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 4.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 4.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 14.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 29.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 19.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 2.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 13.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 6.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 22.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 7.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 9.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.3 | 18.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 2.2 | 12.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.4 | 5.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.1 | 29.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 1.1 | 7.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.0 | 3.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.8 | 5.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.8 | 24.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.8 | 5.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 4.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.7 | 2.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 7.0 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.7 | 2.7 | GO:0019002 | GMP binding(GO:0019002) |
| 0.7 | 4.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.6 | 6.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 3.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.5 | 4.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 6.7 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.5 | 2.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 11.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 1.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.4 | 7.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 1.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.4 | 1.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.4 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 2.6 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.4 | 2.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 3.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 3.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 5.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 3.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 6.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 1.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 4.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 1.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 3.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 3.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 2.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 57.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 7.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 3.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.6 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 12.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.6 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 1.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 4.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 6.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 1.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 7.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 2.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 2.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 9.3 | GO:0046873 | metal ion transmembrane transporter activity(GO:0046873) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 9.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 24.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 27.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 6.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 5.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 11.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 11.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 29.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 7.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 7.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 4.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 17.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 3.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 3.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 5.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 5.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 17.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 4.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.1 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |