Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Gsc2_Dmbx1
Z-value: 0.38
Transcription factors associated with Gsc2_Dmbx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
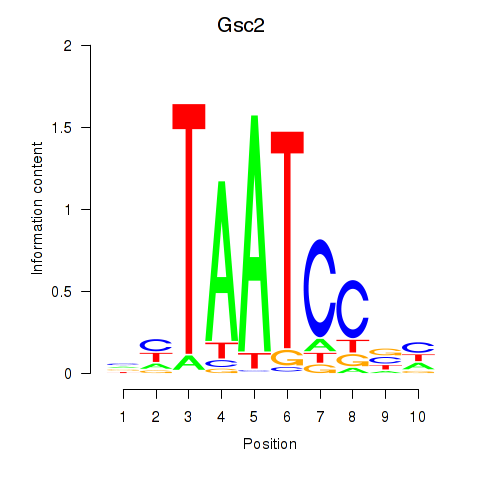
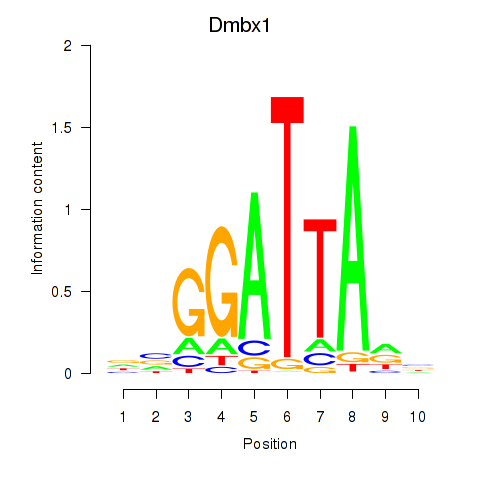
Gsc2
|
ENSRNOG00000000282 | goosecoid homeobox 2 |
|
Dmbx1
|
ENSRNOG00000010835 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmbx1 | rn6_v1_chr5_-_134776247_134776247 | -0.34 | 6.0e-10 | Click! |
| Gsc2 | rn6_v1_chr11_+_87220618_87220618 | 0.19 | 6.9e-04 | Click! |
Activity profile of Gsc2_Dmbx1 motif
Sorted Z-values of Gsc2_Dmbx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_80075991 | 11.85 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr10_+_104368247 | 8.87 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr7_-_143497108 | 7.73 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr15_+_83707735 | 7.09 |
ENSRNOT00000057843
|
Klf5
|
Kruppel-like factor 5 |
| chr3_+_43255567 | 6.59 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr3_+_112242270 | 6.14 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr7_-_49741540 | 5.67 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr1_+_124983452 | 5.06 |
ENSRNOT00000021262
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_-_115969304 | 4.96 |
ENSRNOT00000007958
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chr4_+_151298548 | 4.74 |
ENSRNOT00000010746
|
Cacna2d4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4 |
| chr7_+_94795214 | 4.53 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr4_-_30276372 | 4.33 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr7_-_115969064 | 4.22 |
ENSRNOT00000075999
|
Lypd2
|
Ly6/Plaur domain containing 2 |
| chr4_+_65801505 | 3.55 |
ENSRNOT00000018331
|
Tmem213
|
transmembrane protein 213 |
| chr4_-_125929002 | 3.43 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chrX_-_25628272 | 3.13 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr5_-_135025084 | 3.03 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr8_+_33514042 | 2.93 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr11_+_87220618 | 2.91 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr1_+_99648658 | 2.71 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr2_-_250981623 | 2.40 |
ENSRNOT00000018435
|
Clca2
|
chloride channel accessory 2 |
| chr3_-_113423473 | 2.37 |
ENSRNOT00000064982
|
Serinc4
|
serine incorporator 4 |
| chr1_+_224824799 | 2.34 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr20_-_5155293 | 2.34 |
ENSRNOT00000092322
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr6_+_26780352 | 2.33 |
ENSRNOT00000009916
|
Prr30
|
proline rich 30 |
| chr1_+_219481575 | 2.24 |
ENSRNOT00000025507
|
Pold4
|
DNA polymerase delta 4, accessory subunit |
| chr9_+_51263622 | 2.23 |
ENSRNOT00000004463
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr20_-_8574082 | 2.11 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr8_-_115140080 | 2.08 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr18_-_74542077 | 1.95 |
ENSRNOT00000078519
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr5_+_138300107 | 1.93 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr5_-_115387377 | 1.81 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr7_+_60015998 | 1.79 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr10_-_55560422 | 1.70 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr2_-_250805445 | 1.63 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr20_+_32717564 | 1.62 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr3_-_113423149 | 1.57 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr8_+_55050284 | 1.56 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr1_-_205030567 | 1.48 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
| chrM_+_8599 | 1.35 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr1_-_256734719 | 1.21 |
ENSRNOT00000021546
ENSRNOT00000089456 |
Myof
|
myoferlin |
| chr11_-_69201380 | 1.19 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr8_-_50603591 | 1.16 |
ENSRNOT00000077748
|
LOC500990
|
similar to RIKEN cDNA 4931429L15 |
| chr14_+_85351358 | 1.12 |
ENSRNOT00000031914
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr4_+_117962319 | 1.07 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr7_+_41475163 | 0.98 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr4_+_14001761 | 0.96 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr4_-_29778039 | 0.90 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr3_-_8659102 | 0.88 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr11_-_35697072 | 0.77 |
ENSRNOT00000039999
|
Erg
|
ERG, ETS transcription factor |
| chr19_-_38120578 | 0.77 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chrM_+_7919 | 0.74 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_+_98387291 | 0.70 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr4_-_56656980 | 0.60 |
ENSRNOT00000009278
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr5_-_159577134 | 0.57 |
ENSRNOT00000011114
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr1_+_101412736 | 0.54 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr4_+_28972434 | 0.48 |
ENSRNOT00000014219
|
Gngt1
|
G protein subunit gamma transducin 1 |
| chr10_+_45966926 | 0.46 |
ENSRNOT00000067584
|
Olfr225
|
olfactory receptor 225 |
| chr8_-_114013623 | 0.43 |
ENSRNOT00000018175
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr1_-_199177365 | 0.35 |
ENSRNOT00000041840
|
Ctf2
|
cardiotrophin 2 |
| chr16_+_14319777 | 0.31 |
ENSRNOT00000038771
|
Lrit2
|
leucine-rich repeat, Ig-like and transmembrane domains 2 |
| chr2_+_60966789 | 0.25 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr3_+_73161632 | 0.24 |
ENSRNOT00000077865
|
Olr458
|
olfactory receptor 458 |
| chr4_+_1601652 | 0.17 |
ENSRNOT00000072284
|
LOC100912152
|
olfactory receptor 8B3-like |
| chr5_+_57738321 | 0.16 |
ENSRNOT00000063849
|
Ubap1
|
ubiquitin-associated protein 1 |
| chr2_+_46140482 | 0.16 |
ENSRNOT00000072858
|
Olr1262
|
olfactory receptor 1262 |
| chr5_-_6186329 | 0.08 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr11_+_84827062 | 0.03 |
ENSRNOT00000058006
|
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr16_+_14306804 | 0.03 |
ENSRNOT00000017773
|
Lrit1
|
leucine-rich repeat, Ig-like and transmembrane domains 1 |
| chr10_-_57121584 | 0.03 |
ENSRNOT00000029421
|
Vmo1
|
vitelline membrane outer layer 1 homolog |
| chr4_-_1686845 | 0.02 |
ENSRNOT00000073139
|
LOC100912507
|
olfactory receptor 8B3-like |
| chr16_+_81153489 | 0.00 |
ENSRNOT00000024999
|
Grk1
|
G protein-coupled receptor kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsc2_Dmbx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 2.0 | 6.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 1.4 | 4.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 1.3 | 6.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 1.0 | 2.9 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.7 | 5.7 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.6 | 11.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.6 | 7.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.5 | 7.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.4 | 5.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 2.3 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.4 | 2.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 3.9 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 2.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 4.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.3 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 3.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 4.7 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.7 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.2 | 1.0 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.7 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 1.6 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 1.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 1.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.8 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 1.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 2.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 4.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 3.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.5 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 1.3 | 6.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 4.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 2.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 4.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 6.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 6.2 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 12.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 4.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 1.4 | 4.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.1 | 11.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.7 | 2.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 2.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 6.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 1.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 3.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 2.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 4.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 4.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.2 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 5.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 7.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 5.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 4.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 5.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.9 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 8.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 6.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |