Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
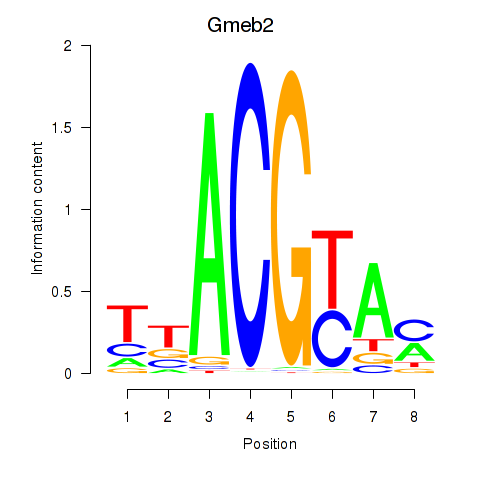
Results for Gmeb2
Z-value: 0.35
Transcription factors associated with Gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gmeb2
|
ENSRNOG00000013339 | glucocorticoid modulatory element binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gmeb2 | rn6_v1_chr3_-_176791960_176791960 | 0.33 | 2.4e-09 | Click! |
Activity profile of Gmeb2 motif
Sorted Z-values of Gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_55001464 | 10.49 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr3_+_171213936 | 10.40 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr7_-_141307233 | 7.07 |
ENSRNOT00000071885
|
Racgap1
|
Rac GTPase-activating protein 1 |
| chr10_-_65424802 | 6.99 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr1_+_220325352 | 6.75 |
ENSRNOT00000027258
|
LOC108348098
|
breast cancer metastasis-suppressor 1 homolog |
| chr1_+_141391262 | 5.81 |
ENSRNOT00000031783
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr7_-_119768082 | 5.72 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chr20_+_2501252 | 5.49 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr17_+_15429708 | 4.59 |
ENSRNOT00000093261
|
LOC679342
|
hypothetical protein LOC679342 |
| chrX_-_15347591 | 4.33 |
ENSRNOT00000037066
|
Timm17b
|
translocase of inner mitochondrial membrane 17b |
| chr14_+_42714315 | 4.23 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr8_+_116054465 | 4.23 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr7_-_14302552 | 4.15 |
ENSRNOT00000091368
|
Brd4
|
bromodomain containing 4 |
| chr9_-_54457753 | 3.26 |
ENSRNOT00000020032
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr5_-_147823232 | 3.21 |
ENSRNOT00000074345
|
Iqcc
|
IQ motif containing C |
| chr3_+_33641616 | 3.16 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr1_+_219345918 | 3.14 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr1_+_167538263 | 3.04 |
ENSRNOT00000074058
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr6_+_52751106 | 2.79 |
ENSRNOT00000014373
|
Twistnb
|
TWIST neighbor |
| chrX_+_159703578 | 2.77 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr11_+_47243342 | 2.75 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_-_85725429 | 2.66 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr17_-_13586554 | 2.61 |
ENSRNOT00000041371
|
Secisbp2
|
SECIS binding protein 2 |
| chr12_-_24724997 | 2.41 |
ENSRNOT00000025560
|
Abhd11
|
abhydrolase domain containing 11 |
| chr8_+_118206698 | 2.40 |
ENSRNOT00000082607
ENSRNOT00000064140 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr6_-_80334522 | 2.38 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr7_-_120780108 | 2.36 |
ENSRNOT00000018185
|
Ddx17
|
DEAD-box helicase 17 |
| chr8_+_104040934 | 2.35 |
ENSRNOT00000081204
|
Tfdp2
|
transcription factor Dp-2 |
| chr6_+_8346704 | 2.33 |
ENSRNOT00000092218
|
AC111831.2
|
|
| chr19_+_37668693 | 2.25 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr8_+_117280705 | 2.25 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr1_+_167538744 | 2.24 |
ENSRNOT00000093070
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr7_-_12399910 | 2.23 |
ENSRNOT00000021677
|
RGD1562114
|
RGD1562114 |
| chr8_+_79660657 | 2.22 |
ENSRNOT00000090970
|
Ccpg1
|
cell cycle progression 1 |
| chr9_+_10428853 | 1.85 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr17_-_14589154 | 1.71 |
ENSRNOT00000073035
|
Nol8
|
nucleolar protein 8 |
| chr1_+_154377447 | 1.65 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_31805728 | 1.63 |
ENSRNOT00000032162
|
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr15_-_19733967 | 1.61 |
ENSRNOT00000012036
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr10_-_56286391 | 1.59 |
ENSRNOT00000018670
|
Senp3
|
Sumo1/sentrin/SMT3 specific peptidase 3 |
| chr19_+_16415636 | 1.59 |
ENSRNOT00000089975
|
Irx5
|
iroquois homeobox 5 |
| chr20_-_27308069 | 1.59 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr7_+_120202601 | 1.58 |
ENSRNOT00000082862
|
AC096473.3
|
|
| chr6_+_96871625 | 1.57 |
ENSRNOT00000012361
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr14_+_84211600 | 1.55 |
ENSRNOT00000005996
|
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr2_+_243422811 | 1.51 |
ENSRNOT00000014694
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr19_+_55300395 | 1.50 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chrX_+_15348138 | 1.48 |
ENSRNOT00000010536
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr1_+_127010588 | 1.40 |
ENSRNOT00000016939
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr5_+_148320438 | 1.39 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr11_-_71419223 | 1.38 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr12_+_11179329 | 1.29 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr14_-_86706626 | 1.22 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr1_-_175895510 | 1.19 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chrX_-_15504165 | 1.16 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr16_+_8207223 | 1.16 |
ENSRNOT00000026751
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr10_+_86724427 | 1.12 |
ENSRNOT00000013204
|
Casc3
|
cancer susceptibility candidate 3 |
| chr1_-_259674425 | 1.10 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chrX_-_105417323 | 1.08 |
ENSRNOT00000015494
|
Gla
|
galactosidase, alpha |
| chr14_+_2100106 | 1.02 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr1_-_259089632 | 1.01 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_-_50943488 | 0.90 |
ENSRNOT00000068419
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr17_-_15429322 | 0.87 |
ENSRNOT00000093381
|
Nol8
|
nucleolar protein 8 |
| chr13_-_73704480 | 0.80 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr7_-_120780641 | 0.77 |
ENSRNOT00000076164
|
Ddx17
|
DEAD-box helicase 17 |
| chr7_-_29282039 | 0.74 |
ENSRNOT00000067777
|
RGD1561102
|
similar to ribosomal protein S12 |
| chr10_+_97647111 | 0.74 |
ENSRNOT00000055062
|
Gna13
|
G protein subunit alpha 13 |
| chr4_-_176789304 | 0.74 |
ENSRNOT00000018057
|
Kcnj8
|
potassium voltage-gated channel subfamily J member 8 |
| chr12_-_39679510 | 0.67 |
ENSRNOT00000001722
|
Gpn3
|
GPN-loop GTPase 3 |
| chr10_-_56289882 | 0.61 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr7_-_70407177 | 0.60 |
ENSRNOT00000049895
|
Os9
|
OS9, endoplasmic reticulum lectin |
| chr4_-_98342887 | 0.58 |
ENSRNOT00000010156
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr18_-_27749235 | 0.55 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr20_+_28027054 | 0.52 |
ENSRNOT00000071386
ENSRNOT00000001044 |
Ranbp2
|
RAN binding protein 2 |
| chr13_-_73704678 | 0.44 |
ENSRNOT00000005280
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr9_+_88110731 | 0.43 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr9_-_98597359 | 0.30 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr8_-_132790778 | 0.26 |
ENSRNOT00000008255
ENSRNOT00000091431 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr20_-_5455632 | 0.21 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr20_-_32080088 | 0.21 |
ENSRNOT00000080773
|
Supv3l1
|
Suv3 like RNA helicase |
| chr5_+_157848206 | 0.20 |
ENSRNOT00000039663
ENSRNOT00000082138 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr13_-_78852160 | 0.16 |
ENSRNOT00000039082
ENSRNOT00000076514 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr3_+_164986421 | 0.10 |
ENSRNOT00000039403
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr1_-_190370499 | 0.07 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr9_+_114022137 | 0.04 |
ENSRNOT00000007657
|
Map3k7
|
mitogen activated protein kinase kinase kinase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.5 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 2.6 | 10.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 2.4 | 7.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.4 | 4.2 | GO:0021550 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 1.0 | 5.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 1.0 | 5.7 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.9 | 2.6 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.8 | 3.3 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.8 | 6.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.7 | 2.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.6 | 7.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.6 | 4.2 | GO:2000002 | negative regulation of cell cycle checkpoint(GO:1901977) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.6 | 1.2 | GO:1905077 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.6 | 1.7 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.5 | 1.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.5 | 1.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.4 | 2.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.4 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 2.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 2.8 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.3 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 5.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 3.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.3 | 1.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 4.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 1.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 1.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.3 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 5.5 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.2 | 2.6 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.2 | 0.7 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 2.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 3.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 0.6 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 2.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 0.6 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 1.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.6 | GO:0007000 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) nucleolus organization(GO:0007000) |
| 0.2 | 1.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 1.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.9 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 1.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.4 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 1.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.9 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.1 | 2.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 4.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.2 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.1 | 0.3 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 1.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.2 | GO:0006342 | chromatin silencing(GO:0006342) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 1.8 | 7.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.3 | 5.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.8 | 3.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.5 | 1.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.5 | 4.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.4 | 4.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.4 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 6.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 2.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 2.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 0.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.5 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.2 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 5.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.2 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.1 | 1.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 10.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.4 | 4.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 1.0 | 5.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.9 | 5.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 5.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.5 | 1.6 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.5 | 10.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 1.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 2.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 2.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 1.6 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.4 | 2.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 3.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 7.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 2.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.3 | 3.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 0.7 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.7 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 7.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 6.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 3.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 4.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 4.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 5.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 11.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 7.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 7.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 5.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 3.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 7.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 4.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 2.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 5.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |