Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
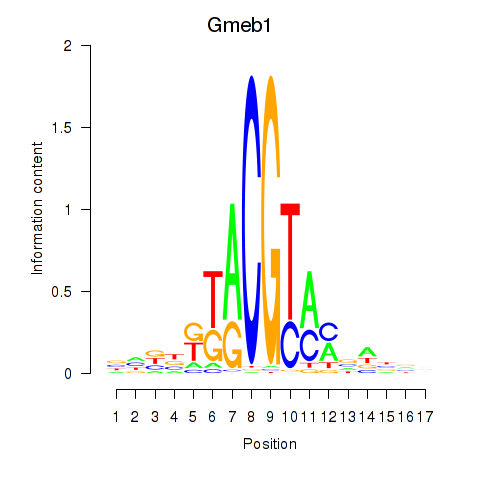
Results for Gmeb1
Z-value: 0.41
Transcription factors associated with Gmeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gmeb1
|
ENSRNOG00000010910 | glucocorticoid modulatory element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gmeb1 | rn6_v1_chr5_-_150439959_150439959 | 0.04 | 4.8e-01 | Click! |
Activity profile of Gmeb1 motif
Sorted Z-values of Gmeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_175709465 | 7.19 |
ENSRNOT00000089971
|
Gata5
|
GATA binding protein 5 |
| chr1_-_222468896 | 6.23 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr10_-_83898527 | 5.34 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr3_+_150801289 | 4.99 |
ENSRNOT00000035060
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr6_-_8346197 | 4.38 |
ENSRNOT00000061826
|
Prepl
|
prolyl endopeptidase-like |
| chr10_-_82252720 | 4.33 |
ENSRNOT00000066132
ENSRNOT00000075795 |
Mycbpap
|
Mycbp associated protein |
| chr17_-_79676499 | 4.32 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chr15_-_37325178 | 4.32 |
ENSRNOT00000011699
|
Gja3
|
gap junction protein, alpha 3 |
| chr10_-_82252963 | 4.05 |
ENSRNOT00000086261
|
Mycbpap
|
Mycbp associated protein |
| chr1_+_261158261 | 3.93 |
ENSRNOT00000071965
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr17_+_11683862 | 3.75 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr1_-_222167447 | 3.72 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr20_-_5155293 | 3.67 |
ENSRNOT00000092322
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr14_-_87465374 | 3.49 |
ENSRNOT00000088355
|
Igfbp3
|
insulin-like growth factor binding protein 3 |
| chr10_+_88914276 | 3.48 |
ENSRNOT00000087076
ENSRNOT00000055238 |
Atp6v0a1
|
ATPase H+ transporting V0 subunit a1 |
| chr1_-_242441247 | 3.39 |
ENSRNOT00000068645
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr9_+_10428853 | 3.23 |
ENSRNOT00000074253
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr12_-_48627297 | 3.17 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr10_+_70520206 | 3.15 |
ENSRNOT00000088198
ENSRNOT00000090446 ENSRNOT00000085799 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr9_-_98597359 | 2.88 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr11_+_33845463 | 2.67 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr7_-_122635731 | 2.20 |
ENSRNOT00000025925
|
St13
|
suppression of tumorigenicity 13 |
| chr16_-_7290561 | 2.18 |
ENSRNOT00000036910
|
Nisch
|
nischarin |
| chr5_+_148320438 | 2.18 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr18_+_31444472 | 2.11 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr12_+_2054680 | 2.11 |
ENSRNOT00000001290
|
Mcoln1
|
mucolipin 1 |
| chr7_+_63922879 | 1.98 |
ENSRNOT00000043581
|
RGD1565498
|
similar to Hypothetical protein LOC270802 |
| chr10_-_39323816 | 1.95 |
ENSRNOT00000081898
ENSRNOT00000041253 |
LOC303140
|
up-regulator of carnitine transporter, OCTN2 |
| chr1_+_90948976 | 1.89 |
ENSRNOT00000056877
|
LOC108348111
|
succinate dehydrogenase assembly factor 1, mitochondrial |
| chr3_+_123754057 | 1.88 |
ENSRNOT00000034201
ENSRNOT00000084671 |
Ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr16_-_74864816 | 1.86 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr7_-_117364697 | 1.77 |
ENSRNOT00000077314
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr1_+_261337594 | 1.75 |
ENSRNOT00000019874
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr8_-_108880879 | 1.74 |
ENSRNOT00000020610
|
Slc35g2
|
solute carrier family 35, member G2 |
| chr1_-_88780425 | 1.69 |
ENSRNOT00000074494
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr1_+_65541322 | 1.53 |
ENSRNOT00000030931
|
Chmp2a
|
charged multivesicular body protein 2A |
| chr9_-_66019065 | 1.53 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chrX_-_15347591 | 1.34 |
ENSRNOT00000037066
|
Timm17b
|
translocase of inner mitochondrial membrane 17b |
| chr7_+_122636171 | 1.27 |
ENSRNOT00000068666
|
Xpnpep3
|
X-prolyl aminopeptidase 3 |
| chr1_-_80221710 | 1.18 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr13_-_73460912 | 1.17 |
ENSRNOT00000005052
ENSRNOT00000068044 |
Qsox1
|
quiescin sulfhydryl oxidase 1 |
| chr14_+_16341536 | 1.08 |
ENSRNOT00000002924
|
Ccni
|
cyclin I |
| chr2_-_231881939 | 1.05 |
ENSRNOT00000074644
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr10_+_63829807 | 1.03 |
ENSRNOT00000006407
|
Crk
|
CRK proto-oncogene, adaptor protein |
| chr18_+_29386809 | 1.01 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr12_+_2461502 | 0.99 |
ENSRNOT00000001415
|
Elavl1
|
ELAV like RNA binding protein 1 |
| chr1_+_100845563 | 0.95 |
ENSRNOT00000027478
|
Akt1s1
|
AKT1 substrate 1 |
| chr1_-_80221417 | 0.92 |
ENSRNOT00000072149
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr10_-_38642397 | 0.85 |
ENSRNOT00000023628
|
Hspa4
|
heat shock protein family A member 4 |
| chr19_+_57771398 | 0.82 |
ENSRNOT00000071827
|
Tsnax
|
translin-associated factor X |
| chr3_-_160038078 | 0.80 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr7_-_120780641 | 0.72 |
ENSRNOT00000076164
|
Ddx17
|
DEAD-box helicase 17 |
| chr10_+_83201311 | 0.56 |
ENSRNOT00000006179
|
Slc35b1
|
solute carrier family 35, member B1 |
| chr2_+_62198562 | 0.54 |
ENSRNOT00000087636
|
Zfr
|
zinc finger RNA binding protein |
| chrX_+_15348138 | 0.53 |
ENSRNOT00000010536
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr15_-_4035064 | 0.52 |
ENSRNOT00000012300
|
Fut11
|
fucosyltransferase 11 |
| chr1_-_261229046 | 0.51 |
ENSRNOT00000075531
|
Mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr3_+_55960067 | 0.48 |
ENSRNOT00000010216
|
Ppig
|
peptidylprolyl isomerase G |
| chr1_-_31545559 | 0.46 |
ENSRNOT00000017779
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr1_-_222734184 | 0.45 |
ENSRNOT00000049812
ENSRNOT00000028785 |
Rtn3
|
reticulon 3 |
| chr7_-_140245723 | 0.38 |
ENSRNOT00000088999
|
Ccnt1
|
cyclin T1 |
| chr7_-_120780108 | 0.33 |
ENSRNOT00000018185
|
Ddx17
|
DEAD-box helicase 17 |
| chr9_+_17041389 | 0.29 |
ENSRNOT00000025598
|
Abcc10
|
ATP binding cassette subfamily C member 10 |
| chr9_+_88110731 | 0.28 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr6_+_8346704 | 0.27 |
ENSRNOT00000092218
|
AC111831.2
|
|
| chr10_-_36716601 | 0.17 |
ENSRNOT00000038838
|
LOC497899
|
similar to hypothetical protein 4930503F14 |
| chr1_-_190370499 | 0.16 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr17_-_1797732 | 0.15 |
ENSRNOT00000033017
ENSRNOT00000079742 |
Cdc14b
|
cell division cycle 14B |
| chr3_-_94657377 | 0.14 |
ENSRNOT00000077484
|
Tcp11l1
|
t-complex 11 like 1 |
| chr9_+_102862890 | 0.14 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr20_-_10257044 | 0.11 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr15_-_83442008 | 0.08 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gmeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.3 | 3.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 1.2 | 3.7 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.0 | 2.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.9 | 3.5 | GO:1902512 | testosterone secretion(GO:0035936) positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) regulation of testosterone secretion(GO:2000843) |
| 0.7 | 2.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.7 | 3.6 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.7 | 2.7 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.6 | 1.8 | GO:0045575 | basophil activation(GO:0045575) |
| 0.6 | 3.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 2.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.5 | 3.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 6.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 3.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 8.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 5.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.3 | 7.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.3 | 1.0 | GO:1990839 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.2 | 3.2 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.2 | 1.5 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.2 | 0.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.6 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.3 | GO:0036093 | germ cell proliferation(GO:0036093) |
| 0.1 | 2.2 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.1 | 3.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 4.3 | GO:0009268 | response to pH(GO:0009268) |
| 0.1 | 1.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.0 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 2.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 2.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 8.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 1.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 1.8 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.6 | 3.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 5.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 6.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 5.0 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 4.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 3.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 4.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 3.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 3.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 3.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 8.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 10.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) gamma-tubulin small complex(GO:0008275) |
| 0.0 | 2.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 3.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 3.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.8 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.6 | 1.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.6 | 1.8 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.6 | 1.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.5 | 2.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.5 | 5.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 3.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.5 | 2.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 2.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.4 | 3.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 2.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 4.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 4.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 5.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 6.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.5 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.1 | 1.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 3.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.8 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 7.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 2.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 3.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 2.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 4.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 3.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 6.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 7.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 4.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |