Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Glis3
Z-value: 0.54
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSRNOG00000014768 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis3 | rn6_v1_chr1_-_246785360_246785360 | -0.01 | 8.4e-01 | Click! |
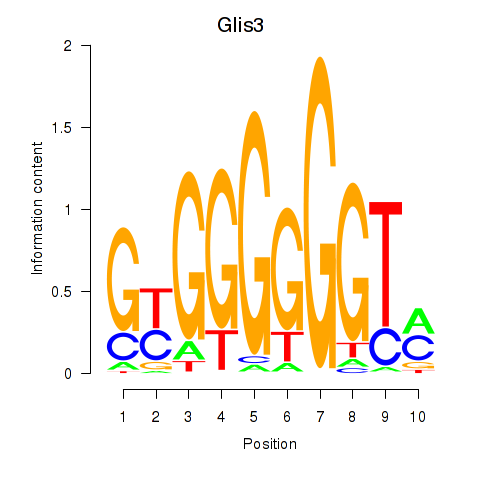
Activity profile of Glis3 motif
Sorted Z-values of Glis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_65275408 | 16.22 |
ENSRNOT00000065137
|
Hmga2
|
high mobility group AT-hook 2 |
| chr3_+_61658245 | 15.61 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr7_+_144014173 | 14.61 |
ENSRNOT00000019403
|
Sp1
|
Sp1 transcription factor |
| chr4_+_82665094 | 12.83 |
ENSRNOT00000078254
|
AABR07060591.1
|
|
| chr5_-_151977636 | 12.43 |
ENSRNOT00000009173
|
Arid1a
|
AT-rich interaction domain 1A |
| chr5_+_162808646 | 12.04 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr10_-_13081136 | 11.70 |
ENSRNOT00000005718
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr5_+_133620706 | 10.95 |
ENSRNOT00000053202
|
Gm12830
|
predicted gene 12830 |
| chr1_+_72636959 | 10.69 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chrX_-_114232939 | 10.24 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr5_-_152247332 | 8.90 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr10_+_89181180 | 8.71 |
ENSRNOT00000078931
ENSRNOT00000027770 |
Wnk4
|
WNK lysine deficient protein kinase 4 |
| chr9_-_92435363 | 8.61 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr12_+_21721837 | 7.55 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr16_-_36373546 | 7.35 |
ENSRNOT00000079552
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr14_+_83510278 | 6.64 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr9_+_25203534 | 6.57 |
ENSRNOT00000064584
|
AABR07066994.1
|
|
| chr10_+_93415399 | 6.42 |
ENSRNOT00000066484
|
Tlk2
|
tousled-like kinase 2 |
| chr14_+_91557601 | 6.37 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr3_-_148312791 | 6.03 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr1_+_243477493 | 5.70 |
ENSRNOT00000021779
ENSRNOT00000085356 |
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr1_+_101012822 | 5.63 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr10_-_64862268 | 5.24 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr1_-_104973648 | 5.21 |
ENSRNOT00000019739
|
Dbx1
|
developing brain homeobox 1 |
| chrX_+_71324365 | 4.97 |
ENSRNOT00000004911
|
Nono
|
non-POU domain containing, octamer-binding |
| chr7_+_144531814 | 4.82 |
ENSRNOT00000033300
|
Hoxc13
|
homeobox C13 |
| chr15_-_45376324 | 4.74 |
ENSRNOT00000076969
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr15_-_45376476 | 4.70 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr1_+_198214797 | 4.45 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr17_-_1093873 | 4.29 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr1_+_225140863 | 4.13 |
ENSRNOT00000027141
|
Mta2
|
metastasis associated 1 family, member 2 |
| chrX_-_63204530 | 3.69 |
ENSRNOT00000076175
|
Zfx
|
zinc finger protein X-linked |
| chr10_+_86711240 | 3.67 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr10_-_83888823 | 3.36 |
ENSRNOT00000055500
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr7_-_140546908 | 3.05 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr3_-_151658417 | 2.75 |
ENSRNOT00000047246
|
LOC100910990
|
copine-1-like |
| chr4_+_140703619 | 2.63 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr20_+_4959294 | 2.59 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr12_-_21760292 | 2.37 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr1_+_154377247 | 2.22 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_78354934 | 2.11 |
ENSRNOT00000020509
|
Zc3h4
|
zinc finger CCCH-type containing 4 |
| chr3_+_11679530 | 2.05 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chrX_+_71155601 | 1.91 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr10_-_88533171 | 1.66 |
ENSRNOT00000024321
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr10_-_85636902 | 1.55 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr1_-_156296161 | 1.44 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr11_-_83926524 | 1.08 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr2_-_195821608 | 0.95 |
ENSRNOT00000028389
ENSRNOT00000071442 |
Snx27
|
sorting nexin family member 27 |
| chr20_+_4371023 | 0.90 |
ENSRNOT00000000500
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_-_38208719 | 0.60 |
ENSRNOT00000019189
|
Kif2a
|
kinesin family member 2A |
| chr1_+_199196059 | 0.26 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr1_-_216080287 | 0.10 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr1_-_80689171 | 0.01 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.6 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 4.1 | 12.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 4.1 | 16.2 | GO:2001038 | senescence-associated heterochromatin focus assembly(GO:0035986) histone H2A phosphorylation(GO:1990164) regulation of cellular response to drug(GO:2001038) |
| 3.9 | 15.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 2.9 | 8.7 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 2.2 | 8.6 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 2.0 | 6.0 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 1.8 | 7.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.4 | 4.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 1.3 | 12.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.3 | 10.7 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 1.3 | 8.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.1 | 5.7 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.1 | 7.6 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.9 | 4.4 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.7 | 2.2 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.7 | 2.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) cardiac jelly development(GO:1905072) |
| 0.6 | 3.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.6 | 4.8 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.6 | 6.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.6 | 5.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.4 | 5.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 1.9 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.2 | 2.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 2.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 1.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 3.7 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 5.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 10.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 3.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 4.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 6.6 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 3.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.4 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 1.6 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.0 | 12.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.9 | 3.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.9 | 6.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.7 | 5.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 4.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 5.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 15.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 3.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 18.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 2.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 8.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 8.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 6.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 10.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 1.4 | 4.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.1 | 8.9 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 1.0 | 6.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.8 | 7.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 8.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.7 | 14.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.5 | 12.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 12.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 2.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 2.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 8.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 5.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 7.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 1.6 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 4.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 5.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 10.7 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 4.4 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 4.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 38.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 3.0 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 20.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 7.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 11.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 4.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 12.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 5.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 6.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 6.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 5.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 12.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 4.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |