Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
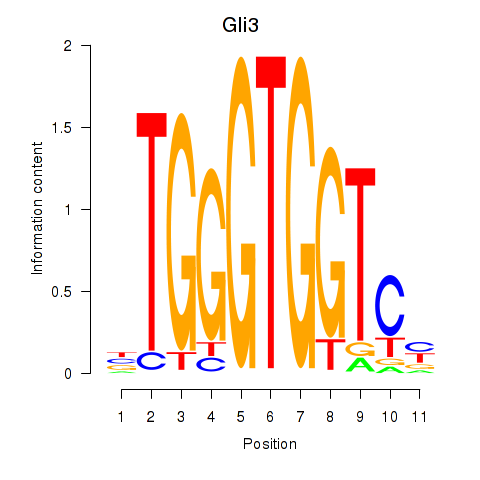
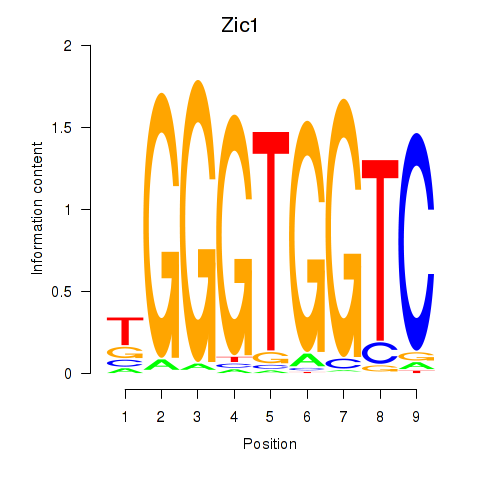
Results for Gli3_Zic1
Z-value: 1.24
Transcription factors associated with Gli3_Zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli3
|
ENSRNOG00000014395 | GLI family zinc finger 3 |
|
Zic1
|
ENSRNOG00000014644 | Zic family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic1 | rn6_v1_chr8_-_98738446_98738446 | 0.67 | 7.4e-43 | Click! |
| Gli3 | rn6_v1_chr17_-_52477575_52477575 | 0.62 | 5.8e-35 | Click! |
Activity profile of Gli3_Zic1 motif
Sorted Z-values of Gli3_Zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_170397191 | 78.31 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chrX_+_71199491 | 69.92 |
ENSRNOT00000076168
ENSRNOT00000005077 ENSRNOT00000005102 |
Nlgn3
|
neuroligin 3 |
| chr10_-_27179254 | 64.87 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr3_+_154043873 | 59.06 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr7_-_136853957 | 58.08 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr10_+_20320878 | 45.80 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr8_+_55178289 | 44.60 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr2_-_29768750 | 43.57 |
ENSRNOT00000023460
|
Map1b
|
microtubule-associated protein 1B |
| chr10_-_14937336 | 40.03 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr2_-_172459165 | 39.00 |
ENSRNOT00000057473
|
Schip1
|
schwannomin interacting protein 1 |
| chr10_+_89089646 | 38.14 |
ENSRNOT00000027505
|
Cntnap1
|
contactin associated protein 1 |
| chr9_+_118842787 | 37.44 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr16_+_54332660 | 35.12 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr7_-_93502571 | 35.05 |
ENSRNOT00000077033
ENSRNOT00000076080 |
Samd12
|
sterile alpha motif domain containing 12 |
| chr5_-_17061361 | 33.18 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr7_-_119441487 | 33.02 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr8_-_59239954 | 32.37 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chrX_-_106747303 | 31.64 |
ENSRNOT00000073529
|
Tceal5
|
transcription elongation factor A like 5 |
| chr4_-_133951264 | 31.08 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr2_-_115836846 | 30.58 |
ENSRNOT00000014359
|
Cldn11
|
claudin 11 |
| chr8_-_55177818 | 30.48 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr1_+_221756286 | 29.70 |
ENSRNOT00000028636
|
Pygm
|
glycogen phosphorylase, muscle associated |
| chr1_+_113034227 | 29.51 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr17_-_18592750 | 29.49 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr7_-_12326392 | 29.46 |
ENSRNOT00000039728
|
Ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7 |
| chrX_+_6791136 | 29.10 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr10_+_86669233 | 28.15 |
ENSRNOT00000012340
|
Thra
|
thyroid hormone receptor alpha |
| chr5_+_140923914 | 27.54 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr4_+_153774486 | 23.05 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr13_+_70379346 | 23.04 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr6_-_27190126 | 22.98 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr7_-_135630654 | 22.83 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr16_-_39476384 | 22.68 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr5_+_18901039 | 22.56 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chrX_+_20216587 | 22.19 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chrX_-_63809861 | 21.60 |
ENSRNOT00000009870
|
Maged1
|
MAGE family member D1 |
| chr1_-_215834704 | 21.29 |
ENSRNOT00000073850
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_216080287 | 21.16 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr11_+_75905443 | 21.11 |
ENSRNOT00000002650
|
Fgf12
|
fibroblast growth factor 12 |
| chr20_-_1980101 | 20.84 |
ENSRNOT00000084582
ENSRNOT00000085050 ENSRNOT00000082545 ENSRNOT00000088396 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr1_-_220467159 | 20.78 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr16_-_39476025 | 20.70 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr1_-_188097530 | 20.22 |
ENSRNOT00000078825
|
Syt17
|
synaptotagmin 17 |
| chr8_+_65686648 | 19.85 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chrX_-_15553720 | 19.82 |
ENSRNOT00000012646
|
Gripap1
|
GRIP1 associated protein 1 |
| chr10_+_91710495 | 19.67 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr8_-_49045154 | 19.34 |
ENSRNOT00000088034
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr6_+_137997335 | 19.16 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr16_+_21288876 | 18.99 |
ENSRNOT00000027990
|
Cilp2
|
cartilage intermediate layer protein 2 |
| chr8_+_119030875 | 18.59 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr12_+_43940929 | 18.44 |
ENSRNOT00000001486
|
Rnft2
|
ring finger protein, transmembrane 2 |
| chr5_+_140870140 | 18.24 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr20_+_3830376 | 18.20 |
ENSRNOT00000085547
|
Col11a2
|
collagen type XI alpha 2 chain |
| chr4_-_145487426 | 17.94 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr8_+_44136496 | 17.71 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr5_-_65073012 | 17.48 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr8_+_119566509 | 17.28 |
ENSRNOT00000028633
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr6_+_27768943 | 17.00 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr3_+_14482388 | 16.58 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr1_-_176607466 | 16.44 |
ENSRNOT00000022984
|
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr9_-_114619711 | 15.99 |
ENSRNOT00000050012
|
Mtcl1
|
microtubule crosslinking factor 1 |
| chr3_+_11593655 | 15.74 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr5_+_135962911 | 15.71 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr1_-_212548730 | 15.48 |
ENSRNOT00000089729
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr20_+_25990656 | 14.95 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr12_+_2534212 | 14.92 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr5_-_85123829 | 14.76 |
ENSRNOT00000007578
|
Brinp1
|
BMP/retinoic acid inducible neural specific 1 |
| chr11_+_87435185 | 14.59 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr1_-_216971183 | 14.55 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr8_-_116361343 | 14.34 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr18_+_83471342 | 14.30 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr12_+_28212333 | 14.12 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr1_+_101783621 | 13.99 |
ENSRNOT00000067679
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr3_+_146582752 | 13.78 |
ENSRNOT00000010157
|
Pygb
|
glycogen phosphorylase B |
| chr2_+_27107318 | 13.76 |
ENSRNOT00000019553
|
Arhgef26
|
Rho guanine nucleotide exchange factor 26 |
| chr1_+_97632473 | 13.59 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr10_-_58693754 | 13.53 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chr1_+_219964429 | 13.49 |
ENSRNOT00000088288
|
Sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr10_-_110585376 | 13.32 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr1_-_80689171 | 12.90 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr9_-_28973246 | 12.71 |
ENSRNOT00000091865
ENSRNOT00000015453 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_18931477 | 12.39 |
ENSRNOT00000077087
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr6_-_122545899 | 12.30 |
ENSRNOT00000005175
|
Kcnk10
|
potassium two pore domain channel subfamily K member 10 |
| chr18_+_29999290 | 12.09 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_+_65448950 | 12.05 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr18_+_58270410 | 12.01 |
ENSRNOT00000067554
|
Apcdd1
|
APC down-regulated 1 |
| chr6_+_99433550 | 12.00 |
ENSRNOT00000079359
ENSRNOT00000008504 |
Hspa2
|
heat shock protein family A member 2 |
| chr20_+_25990304 | 11.95 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr11_+_73738433 | 11.59 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr20_+_3830164 | 11.53 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr19_+_25526751 | 11.24 |
ENSRNOT00000083448
|
Cacna1a
|
calcium voltage-gated channel subunit alpha1 A |
| chr1_+_222519615 | 11.08 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr13_+_80517536 | 10.67 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr15_+_108608204 | 10.65 |
ENSRNOT00000018918
|
Clybl
|
citrate lyase beta like |
| chr9_+_80118029 | 10.58 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr9_+_72052966 | 10.25 |
ENSRNOT00000021099
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr5_-_57896475 | 10.24 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr8_+_129205931 | 10.19 |
ENSRNOT00000082377
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr2_+_195719543 | 10.17 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr13_+_52889737 | 9.73 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr1_-_188097374 | 9.49 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chr9_-_28972835 | 9.45 |
ENSRNOT00000086967
ENSRNOT00000079684 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr11_-_30051103 | 9.23 |
ENSRNOT00000046486
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1 |
| chr7_-_58286770 | 9.09 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chr2_-_188553289 | 9.03 |
ENSRNOT00000088822
|
Trim46
|
tripartite motif-containing 46 |
| chr19_+_22281906 | 9.01 |
ENSRNOT00000021711
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_-_124039196 | 8.84 |
ENSRNOT00000051883
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr16_-_22561496 | 8.76 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
| chr12_+_2046472 | 8.72 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr15_+_86243148 | 8.72 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr1_+_144070754 | 8.69 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr11_-_25078740 | 8.66 |
ENSRNOT00000002109
|
Cyyr1
|
cysteine and tyrosine rich 1 |
| chr9_+_100830250 | 8.49 |
ENSRNOT00000024526
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr12_+_45727112 | 8.42 |
ENSRNOT00000001507
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chr1_-_24056373 | 8.35 |
ENSRNOT00000015566
|
Slc2a12
|
solute carrier family 2 member 12 |
| chr11_-_25350974 | 8.31 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr10_-_105628091 | 8.29 |
ENSRNOT00000016067
|
Cygb
|
cytoglobin |
| chr3_+_173953869 | 8.12 |
ENSRNOT00000091212
|
Fam217b
|
family with sequence similarity 217, member B |
| chr3_-_173953684 | 7.99 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr15_+_40665041 | 7.91 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr3_+_146980923 | 7.77 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr7_+_11769400 | 7.71 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chrX_-_112328642 | 7.58 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr1_-_197886759 | 7.53 |
ENSRNOT00000074274
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr15_-_27819376 | 7.52 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr10_+_69737328 | 7.31 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr17_-_84614228 | 7.14 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr3_+_172385672 | 6.98 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr10_+_36741434 | 6.94 |
ENSRNOT00000064078
|
Col23a1
|
collagen type XXIII alpha 1 chain |
| chr8_-_36410612 | 6.91 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr1_-_49844547 | 6.78 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr6_+_28663602 | 6.74 |
ENSRNOT00000005402
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr1_-_220878059 | 6.70 |
ENSRNOT00000036745
|
Snx32
|
sorting nexin 32 |
| chr12_+_13323547 | 6.69 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr13_-_42263024 | 6.52 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr17_-_10575203 | 6.47 |
ENSRNOT00000073186
|
Arl10
|
ADP-ribosylation factor like GTPase 10 |
| chr19_+_60017746 | 6.34 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr12_-_47095438 | 6.34 |
ENSRNOT00000001546
|
Coq5
|
coenzyme Q5, methyltransferase |
| chr12_+_23752844 | 6.34 |
ENSRNOT00000001953
|
Ssc4d
|
scavenger receptor cysteine rich family member with 4 domains |
| chr18_-_28454756 | 6.30 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr1_-_84986581 | 6.21 |
ENSRNOT00000025819
|
Psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr20_+_21316826 | 6.20 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chrX_-_43592200 | 6.19 |
ENSRNOT00000005033
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr20_-_1984737 | 6.15 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr1_+_199664173 | 6.13 |
ENSRNOT00000054980
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_+_79899155 | 6.03 |
ENSRNOT00000078503
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr11_-_24641820 | 5.98 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr12_-_43940798 | 5.97 |
ENSRNOT00000001485
|
RGD1562310
|
similar to hypothetical protein FLJ21415 |
| chrX_+_119030419 | 5.88 |
ENSRNOT00000060168
|
Pls3
|
plastin 3 |
| chr4_+_140247313 | 5.85 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_+_37305248 | 5.83 |
ENSRNOT00000068299
|
Slc9a5
|
solute carrier family 9 member A5 |
| chr19_-_22281778 | 5.77 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_-_31545559 | 5.77 |
ENSRNOT00000017779
|
Ccdc127
|
coiled-coil domain containing 127 |
| chr19_-_32528965 | 5.74 |
ENSRNOT00000015929
|
Zfp827
|
zinc finger protein 827 |
| chr10_-_13580821 | 5.66 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr11_-_81379871 | 5.57 |
ENSRNOT00000089294
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr14_-_20816588 | 5.54 |
ENSRNOT00000033806
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr4_+_158243086 | 5.50 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr13_+_48607308 | 5.39 |
ENSRNOT00000063882
|
Slc41a1
|
solute carrier family 41 member 1 |
| chrX_-_14642424 | 5.30 |
ENSRNOT00000004808
|
Dynlt3
|
dynein light chain Tctex-type 3 |
| chr3_+_100770975 | 5.22 |
ENSRNOT00000089233
|
Bdnf
|
brain-derived neurotrophic factor |
| chr9_-_82699551 | 5.20 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr8_-_36760742 | 5.15 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr20_+_12773427 | 4.98 |
ENSRNOT00000001695
|
Col6a2
|
collagen type VI alpha 2 chain |
| chr5_-_74190991 | 4.96 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr4_+_71588974 | 4.96 |
ENSRNOT00000037194
|
Tas2r144
|
taste receptor, type 2, member 144 |
| chr3_+_79823945 | 4.90 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr10_+_67677071 | 4.84 |
ENSRNOT00000007312
|
Rhbdl3
|
rhomboid like 3 |
| chr10_+_15099009 | 4.80 |
ENSRNOT00000015206
|
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr6_+_44009872 | 4.73 |
ENSRNOT00000082657
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr19_+_25946979 | 4.68 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr13_+_52147555 | 4.68 |
ENSRNOT00000084766
|
Lmod1
|
leiomodin 1 |
| chr16_-_82439441 | 4.66 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr7_+_12247498 | 4.65 |
ENSRNOT00000022358
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr17_-_1093873 | 4.57 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr12_+_19231092 | 4.56 |
ENSRNOT00000045379
|
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr8_+_3540361 | 4.55 |
ENSRNOT00000050899
|
Vom1r25
|
vomeronasal 1 receptor 25 |
| chr1_+_221099998 | 4.48 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr9_-_16643182 | 4.43 |
ENSRNOT00000024266
|
LOC108348250
|
cullin-7 |
| chr9_-_10734073 | 4.40 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
| chr16_+_80826681 | 4.27 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr9_+_82556573 | 4.21 |
ENSRNOT00000026860
|
Des
|
desmin |
| chrX_-_104337203 | 4.16 |
ENSRNOT00000050903
|
AABR07040565.1
|
|
| chr16_+_22979444 | 4.11 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr5_-_819326 | 4.11 |
ENSRNOT00000023899
|
Pi15
|
peptidase inhibitor 15 |
| chr19_-_56633633 | 4.10 |
ENSRNOT00000023911
|
Ccsap
|
centriole, cilia and spindle-associated protein |
| chr19_-_26141111 | 4.09 |
ENSRNOT00000067518
|
Asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr17_-_10208360 | 4.08 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr9_+_94178221 | 4.07 |
ENSRNOT00000033487
|
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr17_-_21353134 | 3.87 |
ENSRNOT00000067898
|
Smim13
|
small integral membrane protein 13 |
| chr1_-_1767618 | 3.85 |
ENSRNOT00000019623
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr8_+_122003916 | 3.67 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr5_-_77945016 | 3.66 |
ENSRNOT00000076223
|
Zfp37
|
zinc finger protein 37 |
| chr5_-_40867023 | 3.64 |
ENSRNOT00000011576
|
Manea
|
mannosidase, endo-alpha |
| chr3_+_172719432 | 3.61 |
ENSRNOT00000033863
|
Zfp831
|
zinc finger protein 831 |
| chr19_+_54245950 | 3.60 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr2_+_12102487 | 3.55 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr12_+_9034308 | 3.33 |
ENSRNOT00000001248
|
Flt1
|
FMS-related tyrosine kinase 1 |
| chr7_-_96464049 | 3.29 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr11_+_61605937 | 3.26 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli3_Zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 45.8 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 11.1 | 33.2 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 10.0 | 40.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) renal vesicle induction(GO:0072034) |
| 10.0 | 69.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 9.4 | 28.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 7.7 | 23.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 7.6 | 30.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 7.4 | 22.2 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 6.7 | 94.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 6.6 | 19.8 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 6.5 | 32.4 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 5.6 | 44.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 5.5 | 16.6 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 5.3 | 21.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 5.0 | 29.7 | GO:0060023 | soft palate development(GO:0060023) |
| 4.8 | 38.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 4.8 | 14.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 4.7 | 14.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 4.7 | 37.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 4.5 | 27.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 4.2 | 21.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 3.7 | 11.2 | GO:0050883 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 3.5 | 56.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 3.4 | 27.5 | GO:0072014 | proximal tubule development(GO:0072014) |
| 3.4 | 43.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 3.2 | 16.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 3.0 | 21.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 3.0 | 12.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 2.9 | 25.7 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 2.8 | 8.5 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 2.7 | 32.5 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 2.7 | 10.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) ERBB3 signaling pathway(GO:0038129) |
| 2.5 | 17.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 2.5 | 7.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 2.3 | 51.5 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 2.3 | 7.0 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 2.3 | 9.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 2.2 | 30.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 2.0 | 21.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.9 | 5.8 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 1.8 | 36.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 1.8 | 39.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 1.8 | 23.0 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 1.7 | 8.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 1.5 | 23.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.5 | 17.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.5 | 4.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.4 | 78.3 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 1.4 | 9.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.4 | 4.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.3 | 12.0 | GO:0090385 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 1.3 | 8.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.2 | 14.8 | GO:0007614 | short-term memory(GO:0007614) |
| 1.2 | 13.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.2 | 6.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.2 | 35.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 1.1 | 10.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 1.1 | 9.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 1.1 | 11.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 1.1 | 3.3 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 1.1 | 7.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.1 | 8.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.1 | 43.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 1.1 | 13.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 1.1 | 6.3 | GO:0003383 | apical constriction(GO:0003383) |
| 1.0 | 7.3 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 1.0 | 9.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 1.0 | 28.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 1.0 | 17.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.9 | 2.8 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.9 | 8.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.9 | 15.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.9 | 19.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.9 | 4.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.8 | 5.8 | GO:0050882 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) voluntary musculoskeletal movement(GO:0050882) |
| 0.8 | 2.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 2.2 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.7 | 4.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.7 | 7.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.6 | 5.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.6 | 31.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.6 | 4.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.6 | 1.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.6 | 18.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.6 | 4.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 45.7 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.6 | 3.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.6 | 7.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 5.8 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 2.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.5 | 4.7 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.5 | 5.9 | GO:0051764 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.4 | 10.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 5.9 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.4 | 7.7 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.4 | 2.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 6.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 13.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.4 | 6.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.4 | 19.9 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.4 | 1.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.4 | 2.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 3.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 1.0 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 3.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 3.8 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.3 | 4.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.3 | 0.6 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) |
| 0.3 | 2.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 2.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.3 | 8.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 6.5 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 14.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.2 | 5.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 10.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 4.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 2.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 2.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 4.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 6.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 4.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 5.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.2 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 2.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 7.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 10.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 5.8 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 2.9 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.1 | 12.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 14.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 6.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.6 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.1 | 10.9 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.1 | 5.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 2.9 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 5.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 1.8 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 4.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.1 | 84.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 9.9 | 29.7 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 9.0 | 27.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 6.6 | 19.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 5.5 | 33.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 5.4 | 48.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 4.8 | 186.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 4.0 | 16.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 4.0 | 19.8 | GO:0043293 | apoptosome(GO:0043293) |
| 3.6 | 10.7 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 3.0 | 9.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 2.8 | 16.6 | GO:0030478 | actin cap(GO:0030478) |
| 2.6 | 7.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 2.4 | 17.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 2.2 | 13.5 | GO:0008091 | spectrin(GO:0008091) |
| 2.2 | 43.6 | GO:0043196 | varicosity(GO:0043196) |
| 2.1 | 18.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.8 | 9.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.7 | 12.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.6 | 9.6 | GO:1990393 | 3M complex(GO:1990393) |
| 1.5 | 23.0 | GO:0045180 | basal cortex(GO:0045180) |
| 1.5 | 38.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.5 | 64.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.5 | 5.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.4 | 13.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 1.4 | 15.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.4 | 4.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 1.3 | 17.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.3 | 14.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.0 | 17.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 10.2 | GO:0002177 | manchette(GO:0002177) |
| 0.9 | 3.6 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.8 | 8.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.8 | 2.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.8 | 47.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.7 | 5.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 36.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.7 | 2.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.6 | 2.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 15.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.5 | 29.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.5 | 31.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 30.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.5 | 18.6 | GO:0031672 | A band(GO:0031672) |
| 0.5 | 6.1 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.4 | 12.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 71.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 52.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.3 | 8.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 24.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 4.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 5.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 3.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 55.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 1.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 2.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 4.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 4.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 15.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 11.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 24.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 5.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 8.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 11.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 17.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 107.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 1.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 11.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 3.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 9.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 13.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 10.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 43.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 9.2 | 27.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 7.7 | 23.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 7.4 | 51.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 6.7 | 27.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 5.5 | 33.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 5.0 | 69.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 4.9 | 29.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 4.6 | 64.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 4.6 | 23.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 4.1 | 16.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 3.2 | 68.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 3.0 | 21.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 2.9 | 23.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 2.9 | 37.4 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 2.8 | 28.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 2.8 | 8.3 | GO:0004096 | catalase activity(GO:0004096) |
| 2.6 | 15.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 2.6 | 28.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 2.5 | 55.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.3 | 2.3 | GO:0016635 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 2.2 | 17.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.2 | 21.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.1 | 10.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.0 | 15.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.9 | 11.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.9 | 5.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.8 | 35.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.8 | 10.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.7 | 17.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.7 | 6.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.5 | 6.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.5 | 10.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.5 | 6.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.5 | 5.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.4 | 4.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 1.3 | 28.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.3 | 10.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.3 | 29.5 | GO:0048038 | quinone binding(GO:0048038) |
| 1.1 | 21.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 6.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.0 | 64.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.0 | 3.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.8 | 4.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.7 | 5.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 12.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.7 | 2.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.7 | 2.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 21.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.7 | 3.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 5.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 3.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.6 | 18.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.6 | 10.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.6 | 5.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 8.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.5 | 8.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.5 | 12.0 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.5 | 5.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 10.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 29.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.5 | 3.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.5 | 29.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) clathrin binding(GO:0030276) |
| 0.5 | 1.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.5 | 18.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.5 | 6.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 8.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 39.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 14.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.4 | 9.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 5.4 | GO:0015491 | cation:cation antiporter activity(GO:0015491) |
| 0.4 | 12.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 14.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.4 | 35.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.4 | 5.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 3.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 17.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 35.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 4.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 4.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 2.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 24.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.3 | 3.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 5.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 4.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 3.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 2.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 1.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 4.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 3.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.6 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 1.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 7.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 20.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 10.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 32.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 30.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 17.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 6.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 9.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 7.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 5.0 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 31.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 5.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.2 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 8.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 5.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 6.6 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 1.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 11.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 4.4 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 64.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 1.1 | 21.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.9 | 78.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.9 | 41.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.8 | 9.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.6 | 132.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.6 | 16.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.5 | 18.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 26.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 3.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 18.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 5.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 17.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 13.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 11.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 94.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 2.7 | 35.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 2.5 | 49.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.8 | 25.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.6 | 22.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.5 | 31.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.3 | 21.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 1.2 | 27.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.1 | 36.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.9 | 31.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 44.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.7 | 29.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.7 | 41.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.7 | 16.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 22.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 12.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 35.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 38.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.5 | 6.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 8.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 9.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.5 | 7.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 24.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.4 | 5.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.4 | 23.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 38.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.4 | 3.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 2.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 4.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 5.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 16.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 6.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 4.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 5.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 11.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 3.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 9.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 3.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 8.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 22.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 8.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 4.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |