Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
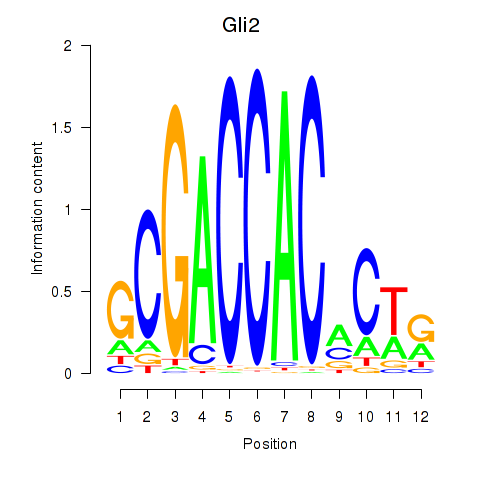
Results for Gli2
Z-value: 0.74
Transcription factors associated with Gli2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli2
|
ENSRNOG00000007261 | GLI family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli2 | rn6_v1_chr13_-_35048444_35048444 | -0.13 | 1.8e-02 | Click! |
Activity profile of Gli2 motif
Sorted Z-values of Gli2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_170431073 | 25.60 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr8_+_116332796 | 21.15 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr1_-_85220237 | 17.82 |
ENSRNOT00000026907
|
Sycn
|
syncollin |
| chr3_+_170354141 | 16.95 |
ENSRNOT00000005901
|
Fam210b
|
family with sequence similarity 210, member B |
| chr1_-_256813711 | 16.85 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr7_+_54980120 | 14.82 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr20_-_2652952 | 14.24 |
ENSRNOT00000078871
|
C4a
|
complement component 4A (Rodgers blood group) |
| chr7_+_120125633 | 13.96 |
ENSRNOT00000012480
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr4_-_78458179 | 13.34 |
ENSRNOT00000078473
ENSRNOT00000011327 |
Tmem176b
|
transmembrane protein 176B |
| chr6_-_92643847 | 12.87 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chr1_-_146736261 | 10.45 |
ENSRNOT00000068167
|
Fah
|
fumarylacetoacetate hydrolase |
| chr17_-_90756048 | 9.44 |
ENSRNOT00000085669
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr3_+_61658245 | 9.44 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr8_-_25904564 | 9.20 |
ENSRNOT00000082744
ENSRNOT00000064783 |
Tbx20
|
T-box 20 |
| chr3_-_123630929 | 8.57 |
ENSRNOT00000028855
|
Siglec1
|
sialic acid binding Ig like lectin 1 |
| chr8_+_49354115 | 8.48 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr2_-_149417212 | 8.08 |
ENSRNOT00000018573
|
Gpr87
|
G protein-coupled receptor 87 |
| chr17_-_90755852 | 7.69 |
ENSRNOT00000003526
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr3_-_83048289 | 7.48 |
ENSRNOT00000047571
ENSRNOT00000012806 |
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr10_-_13580821 | 7.26 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr6_+_12253788 | 7.19 |
ENSRNOT00000061675
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr12_+_30165694 | 7.17 |
ENSRNOT00000001211
|
Asl
|
argininosuccinate lyase |
| chr5_+_135962911 | 6.66 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr16_+_23668595 | 6.57 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_80351774 | 6.30 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chrX_+_70563570 | 5.75 |
ENSRNOT00000003772
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_-_242441247 | 5.71 |
ENSRNOT00000068645
|
Pip5k1b
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr6_+_58468155 | 5.69 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr14_+_83341851 | 5.64 |
ENSRNOT00000086090
|
Pisd
|
phosphatidylserine decarboxylase |
| chr1_-_281756159 | 5.53 |
ENSRNOT00000013170
|
Prlhr
|
prolactin releasing hormone receptor |
| chr4_-_100660140 | 5.44 |
ENSRNOT00000020005
|
Tcf7l1
|
transcription factor 7 like 1 |
| chr8_-_47529689 | 5.07 |
ENSRNOT00000012826
|
Oaf
|
out at first homolog |
| chr8_-_22821397 | 5.05 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr6_+_126170911 | 4.97 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr4_-_132740938 | 4.94 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr8_-_22821223 | 4.88 |
ENSRNOT00000014135
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr6_+_126170720 | 4.79 |
ENSRNOT00000065246
|
Rin3
|
Ras and Rab interactor 3 |
| chr2_-_196526886 | 4.68 |
ENSRNOT00000077325
|
Setdb1
|
SET domain, bifurcated 1 |
| chr9_+_54558202 | 4.67 |
ENSRNOT00000068433
|
Myo1b
|
myosin Ib |
| chr2_+_125752130 | 4.64 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr6_-_51257625 | 4.61 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr12_-_44999074 | 4.40 |
ENSRNOT00000079177
ENSRNOT00000038293 |
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr16_-_9430743 | 4.39 |
ENSRNOT00000043811
|
Wdfy4
|
WDFY family member 4 |
| chr12_+_41385241 | 4.33 |
ENSRNOT00000074974
|
Dtx1
|
deltex E3 ubiquitin ligase 1 |
| chr2_-_196527127 | 4.32 |
ENSRNOT00000028709
|
Setdb1
|
SET domain, bifurcated 1 |
| chr4_+_171748273 | 4.27 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr1_+_221099998 | 4.16 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr10_+_65448950 | 4.14 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr6_-_7421456 | 4.05 |
ENSRNOT00000006725
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chrX_+_20216587 | 3.94 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr1_-_52544450 | 3.93 |
ENSRNOT00000043474
|
Pde10a
|
phosphodiesterase 10A |
| chr19_+_52086325 | 3.83 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr14_+_60123169 | 3.81 |
ENSRNOT00000006610
|
Sel1l3
|
SEL1L family member 3 |
| chr1_+_215214853 | 3.58 |
ENSRNOT00000071235
ENSRNOT00000080442 |
LOC103690160
|
SH3 and multiple ankyrin repeat domains protein 2-like |
| chr10_+_103934797 | 3.53 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr13_+_85580828 | 3.35 |
ENSRNOT00000005611
|
Aldh9a1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr10_+_58860940 | 3.28 |
ENSRNOT00000056551
ENSRNOT00000074523 |
XAF1
|
XIAP associated factor-1 |
| chr4_-_82702429 | 3.25 |
ENSRNOT00000011069
|
Hibadh
|
3-hydroxyisobutyrate dehydrogenase |
| chr2_+_248249468 | 3.20 |
ENSRNOT00000022648
|
Gbp4
|
guanylate binding protein 4 |
| chr3_-_11410732 | 3.18 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr20_+_5933303 | 3.07 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr1_-_190965115 | 3.05 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr7_-_119716238 | 2.93 |
ENSRNOT00000075678
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr2_+_189430041 | 2.76 |
ENSRNOT00000023567
ENSRNOT00000023605 |
Tpm3
|
tropomyosin 3 |
| chr17_+_56935451 | 2.70 |
ENSRNOT00000058966
|
RGD1564129
|
similar to hypothetical protein 4930474N05 |
| chr1_+_190964885 | 2.55 |
ENSRNOT00000039186
|
Mettl9
|
methyltransferase like 9 |
| chr8_-_122841477 | 2.51 |
ENSRNOT00000014861
|
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_+_141767940 | 2.37 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr10_-_71441389 | 2.31 |
ENSRNOT00000003699
|
Tada2a
|
transcriptional adaptor 2A |
| chr10_+_104582955 | 2.30 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr7_-_11443357 | 2.27 |
ENSRNOT00000082126
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr8_-_59849427 | 2.20 |
ENSRNOT00000020332
ENSRNOT00000083900 |
Nrg4
|
neuregulin 4 |
| chr4_-_50312608 | 1.88 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr1_-_214511529 | 1.85 |
ENSRNOT00000026390
|
Chid1
|
chitinase domain containing 1 |
| chr7_-_2353875 | 1.80 |
ENSRNOT00000074873
|
LOC100911319
|
zinc finger protein 36, C3H1 type-like 2-like |
| chr7_+_2718700 | 1.79 |
ENSRNOT00000050792
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr4_+_168976859 | 1.79 |
ENSRNOT00000068663
|
Fam234b
|
family with sequence similarity 234, member B |
| chr10_-_90265017 | 1.71 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr18_-_28017925 | 1.66 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chrX_+_53360839 | 1.65 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr10_+_107502695 | 1.60 |
ENSRNOT00000038088
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr15_-_60056582 | 1.57 |
ENSRNOT00000087569
ENSRNOT00000012530 |
Dnajc15
|
DnaJ heat shock protein family (Hsp40) member C15 |
| chr20_-_4401610 | 1.55 |
ENSRNOT00000091468
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_+_57462447 | 1.54 |
ENSRNOT00000041443
|
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr9_-_100624638 | 1.52 |
ENSRNOT00000051155
|
Hdlbp
|
high density lipoprotein binding protein |
| chr9_-_43022998 | 1.43 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr9_-_11108741 | 1.42 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr8_-_107380933 | 1.42 |
ENSRNOT00000022179
|
Pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr5_-_77031120 | 1.37 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr9_+_111220858 | 1.36 |
ENSRNOT00000076669
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr17_+_57864244 | 1.34 |
ENSRNOT00000089527
|
LOC102551575
|
isopentenyl-diphosphate delta-isomerase 2-like |
| chr1_+_81456984 | 1.29 |
ENSRNOT00000027075
|
Ethe1
|
ETHE1, persulfide dioxygenase |
| chr1_-_214074663 | 1.25 |
ENSRNOT00000026215
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr14_-_83641892 | 1.18 |
ENSRNOT00000026032
|
Limk2
|
LIM domain kinase 2 |
| chrX_+_73390903 | 1.08 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chrX_+_28435507 | 1.02 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chrX_+_156863754 | 0.99 |
ENSRNOT00000083611
|
Naa10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr2_+_18937458 | 0.97 |
ENSRNOT00000022377
|
Tmem167a
|
transmembrane protein 167A |
| chr3_+_112371677 | 0.94 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr2_-_94730978 | 0.92 |
ENSRNOT00000087598
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr6_+_132702448 | 0.86 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr7_-_11257977 | 0.85 |
ENSRNOT00000027932
|
Tbxa2r
|
thromboxane A2 receptor |
| chr17_+_47241017 | 0.84 |
ENSRNOT00000092193
|
Gpr141
|
G protein-coupled receptor 141 |
| chr20_+_3555135 | 0.84 |
ENSRNOT00000085380
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr5_-_59198650 | 0.84 |
ENSRNOT00000020958
|
Olr833
|
olfactory receptor 833 |
| chr10_-_57671080 | 0.83 |
ENSRNOT00000082511
|
LOC691995
|
hypothetical protein LOC691995 |
| chr12_+_21721837 | 0.79 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr7_-_70552897 | 0.75 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr19_+_60017746 | 0.66 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr3_-_3691972 | 0.65 |
ENSRNOT00000061735
|
Qsox2
|
quiescin sulfhydryl oxidase 2 |
| chr3_-_165741967 | 0.64 |
ENSRNOT00000017063
|
Zfp64
|
zinc finger protein 64 |
| chr12_-_17972737 | 0.58 |
ENSRNOT00000001783
|
Fam20c
|
FAM20C, golgi associated secretory pathway kinase |
| chr8_+_55279373 | 0.54 |
ENSRNOT00000064290
|
Ppp2r1b
|
protein phosphatase 2 scaffold subunit A beta |
| chr19_+_10519493 | 0.50 |
ENSRNOT00000030229
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr6_-_111572949 | 0.48 |
ENSRNOT00000016884
|
RGD1565693
|
similar to GLE1-like, RNA export mediator isoform 1 |
| chr9_-_15700235 | 0.47 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr10_-_56591364 | 0.43 |
ENSRNOT00000032481
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr17_-_2705123 | 0.41 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr8_+_25246292 | 0.38 |
ENSRNOT00000021625
|
Npsr1
|
neuropeptide S receptor 1 |
| chr1_-_59347472 | 0.34 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr17_-_27452314 | 0.34 |
ENSRNOT00000018936
|
Riok1
|
RIO kinase 1 |
| chr5_+_171441560 | 0.31 |
ENSRNOT00000019873
|
Wrap73
|
WD repeat containing, antisense to TP73 |
| chr15_+_48327461 | 0.30 |
ENSRNOT00000018071
|
Ints9
|
integrator complex subunit 9 |
| chr8_-_116532169 | 0.30 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr4_+_6559545 | 0.26 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chrX_+_29157470 | 0.19 |
ENSRNOT00000081986
|
LOC100910245
|
ribose-phosphate pyrophosphokinase 2-like |
| chr9_+_26841299 | 0.18 |
ENSRNOT00000016664
|
Il17a
|
interleukin 17A |
| chr7_-_9193141 | 0.17 |
ENSRNOT00000046955
|
Olr1061
|
olfactory receptor 1061 |
| chr1_+_198409360 | 0.17 |
ENSRNOT00000013691
ENSRNOT00000091295 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr8_+_32530806 | 0.12 |
ENSRNOT00000083659
|
Nfrkb
|
nuclear factor related to kappa B binding protein |
| chr5_-_144341586 | 0.07 |
ENSRNOT00000014586
|
Adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr19_-_25288335 | 0.02 |
ENSRNOT00000050214
|
Nanos3
|
nanos C2HC-type zinc finger 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 25.6 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 5.6 | 16.8 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 5.3 | 21.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 3.5 | 13.9 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 3.4 | 17.1 | GO:0030070 | insulin processing(GO:0030070) |
| 3.3 | 13.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.3 | 9.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 3.1 | 9.2 | GO:0060577 | visceral motor neuron differentiation(GO:0021524) pulmonary vein morphogenesis(GO:0060577) |
| 2.5 | 14.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.4 | 7.2 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 2.4 | 9.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 2.3 | 9.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.7 | 10.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.5 | 4.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.4 | 4.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.4 | 5.4 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 1.1 | 3.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.9 | 4.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.8 | 7.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.8 | 7.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.8 | 3.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 8.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.6 | 2.5 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.6 | 2.9 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.6 | 1.7 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.6 | 3.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 1.6 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.5 | 5.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 5.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.5 | 8.6 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.5 | 1.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 4.1 | GO:0090385 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 14.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 5.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 1.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 | 2.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 4.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.3 | 1.3 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.3 | 4.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 3.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 6.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 0.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 0.9 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.2 | 1.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 3.2 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.2 | 0.6 | GO:0097187 | odontoblast differentiation(GO:0071895) dentinogenesis(GO:0097187) |
| 0.2 | 1.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.9 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 1.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 4.6 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 4.3 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.1 | 5.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.1 | 0.8 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.4 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 11.7 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 | 1.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 8.5 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 4.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 4.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 2.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 4.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 3.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 1.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.8 | GO:0006936 | muscle contraction(GO:0006936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.8 | 3.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 7.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.4 | 14.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 2.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 2.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 5.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 9.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 17.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 25.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 4.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 5.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) filopodium membrane(GO:0031527) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 4.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 8.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 9.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 20.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 8.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 5.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 22.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 8.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 21.9 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 3.5 | 10.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.8 | 25.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.6 | 13.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 2.5 | 7.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.8 | 7.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.4 | 5.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.3 | 16.8 | GO:0019841 | retinol binding(GO:0019841) |
| 1.1 | 17.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 1.0 | 2.9 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 1.0 | 5.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.7 | 5.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.7 | 8.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.6 | 13.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 3.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.5 | 3.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 5.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.4 | 2.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.4 | 3.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 0.3 | 1.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.3 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 4.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.3 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 9.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 6.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 3.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 4.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 3.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 9.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.7 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 3.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 9.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 4.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 4.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 4.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.9 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.1 | 14.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 5.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 4.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 8.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 4.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 6.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 5.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 25.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 10.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 21.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 21.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.0 | 14.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 12.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 0.9 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.4 | 7.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 3.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.3 | 14.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 7.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 7.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 22.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |