Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
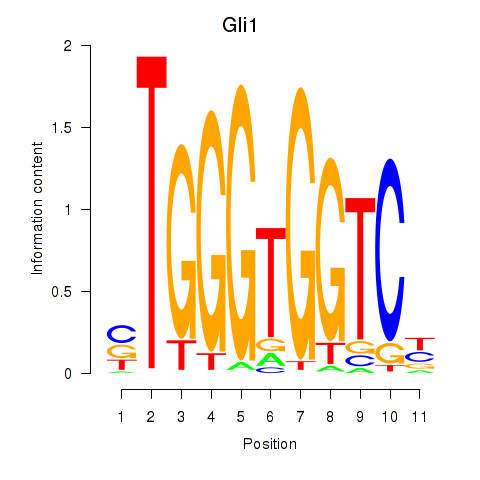
Results for Gli1
Z-value: 0.74
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSRNOG00000025120 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli1 | rn6_v1_chr7_-_70630338_70630338 | 0.49 | 1.3e-20 | Click! |
Activity profile of Gli1 motif
Sorted Z-values of Gli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_39476384 | 31.47 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr16_-_39476025 | 28.05 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr7_-_136853957 | 25.77 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr10_-_27179254 | 23.69 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr3_+_154043873 | 22.44 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr1_-_170397191 | 20.33 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr9_-_75528644 | 20.26 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr11_-_30051103 | 19.72 |
ENSRNOT00000046486
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1 |
| chr6_+_27768943 | 19.69 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr1_-_220467159 | 18.64 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chrX_+_71199491 | 17.82 |
ENSRNOT00000076168
ENSRNOT00000005077 ENSRNOT00000005102 |
Nlgn3
|
neuroligin 3 |
| chr8_-_59239954 | 17.66 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr10_+_89089646 | 17.28 |
ENSRNOT00000027505
|
Cntnap1
|
contactin associated protein 1 |
| chr13_+_70379346 | 17.25 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr10_+_20320878 | 16.79 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr1_+_113034227 | 15.92 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr18_+_79406381 | 14.95 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr1_-_188097530 | 14.75 |
ENSRNOT00000078825
|
Syt17
|
synaptotagmin 17 |
| chr10_+_38889370 | 14.16 |
ENSRNOT00000091236
|
Sept8
|
septin 8 |
| chr16_+_7212488 | 13.31 |
ENSRNOT00000024840
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr5_-_17061361 | 13.26 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr20_-_1984737 | 13.16 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr6_-_4520604 | 11.86 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr10_+_67677071 | 11.64 |
ENSRNOT00000007312
|
Rhbdl3
|
rhomboid like 3 |
| chr10_-_108321644 | 11.29 |
ENSRNOT00000075171
|
Tbc1d16
|
TBC1 domain family, member 16 |
| chr7_+_141326950 | 10.86 |
ENSRNOT00000084075
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr1_-_216080287 | 10.56 |
ENSRNOT00000027682
|
Th
|
tyrosine hydroxylase |
| chr4_+_140247313 | 10.18 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_+_131380297 | 10.14 |
ENSRNOT00000010548
|
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr1_-_84812486 | 9.84 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr1_+_115975324 | 9.38 |
ENSRNOT00000080907
|
Atp10a
|
ATPase phospholipid transporting 10A (putative) |
| chr18_+_58270410 | 8.96 |
ENSRNOT00000067554
|
Apcdd1
|
APC down-regulated 1 |
| chr15_+_40665041 | 8.90 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr6_-_27190126 | 8.76 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr10_+_86669233 | 7.92 |
ENSRNOT00000012340
|
Thra
|
thyroid hormone receptor alpha |
| chr5_+_18901039 | 7.37 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr3_-_39596718 | 7.31 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr17_-_10575203 | 7.16 |
ENSRNOT00000073186
|
Arl10
|
ADP-ribosylation factor like GTPase 10 |
| chr3_+_152752091 | 7.09 |
ENSRNOT00000037177
|
Dlgap4
|
DLG associated protein 4 |
| chr4_+_87293871 | 6.95 |
ENSRNOT00000090943
|
AABR07060628.1
|
|
| chr1_-_188097374 | 6.85 |
ENSRNOT00000092246
|
Syt17
|
synaptotagmin 17 |
| chrX_-_115426083 | 6.73 |
ENSRNOT00000014756
|
AABR07040947.1
|
|
| chr5_-_65073012 | 6.29 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr17_-_1093873 | 6.19 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr19_-_10358695 | 6.09 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr6_+_137997335 | 6.00 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr7_+_14643704 | 5.46 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr20_+_4020317 | 5.42 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chr3_+_80676820 | 5.22 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr3_+_79823945 | 5.19 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr7_-_142300382 | 5.17 |
ENSRNOT00000048262
|
Bin2
|
bridging integrator 2 |
| chr4_+_24617008 | 5.03 |
ENSRNOT00000086607
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr5_+_156618962 | 4.84 |
ENSRNOT00000020069
|
Sh2d5
|
SH2 domain containing 5 |
| chr10_+_15099009 | 4.84 |
ENSRNOT00000015206
|
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chrX_+_28435507 | 4.76 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr1_+_144070754 | 4.65 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr12_+_47074200 | 4.53 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr6_+_86713803 | 4.43 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr10_+_84986328 | 4.41 |
ENSRNOT00000085830
ENSRNOT00000067665 |
Osbpl7
|
oxysterol binding protein-like 7 |
| chr2_-_178057157 | 4.39 |
ENSRNOT00000091135
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_+_77322012 | 4.31 |
ENSRNOT00000088600
ENSRNOT00000041639 |
Zbtb49
|
zinc finger and BTB domain containing 49 |
| chr4_-_145487426 | 4.23 |
ENSRNOT00000013488
|
Emc3
|
ER membrane protein complex subunit 3 |
| chrX_+_29157470 | 4.16 |
ENSRNOT00000081986
|
LOC100910245
|
ribose-phosphate pyrophosphokinase 2-like |
| chr10_+_56824505 | 4.13 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr7_-_12810570 | 3.97 |
ENSRNOT00000012578
|
Fstl3
|
follistatin like 3 |
| chr16_+_61758917 | 3.93 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr6_+_86713604 | 3.79 |
ENSRNOT00000059271
|
Fam179b
|
family with sequence similarity 179, member B |
| chr3_-_2573387 | 3.73 |
ENSRNOT00000017271
|
Dpp7
|
dipeptidylpeptidase 7 |
| chr10_+_56610051 | 3.62 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr2_+_248715281 | 3.51 |
ENSRNOT00000089648
|
Gtf2b
|
general transcription factor IIB |
| chr1_-_210739600 | 3.50 |
ENSRNOT00000031734
|
Tcerg1l
|
transcription elongation regulator 1-like |
| chr10_-_90312386 | 3.34 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr5_+_63781801 | 3.30 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr3_-_147481073 | 3.23 |
ENSRNOT00000084722
|
Fam110a
|
family with sequence similarity 110, member A |
| chr4_+_113935492 | 3.23 |
ENSRNOT00000035329
|
LOC103692170
|
coiled-coil domain-containing protein 142 |
| chr3_-_138708332 | 2.99 |
ENSRNOT00000010678
|
Rbbp9
|
RB binding protein 9, serine hydrolase |
| chr10_+_65448950 | 2.97 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr3_-_148312791 | 2.84 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chrX_-_72370044 | 2.73 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr1_-_80689171 | 2.58 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr19_+_46761570 | 2.43 |
ENSRNOT00000058779
|
Wwox
|
WW domain-containing oxidoreductase |
| chr8_-_23084879 | 2.38 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr13_-_42263024 | 2.27 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chrX_-_43592200 | 2.21 |
ENSRNOT00000005033
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr18_+_60496778 | 2.21 |
ENSRNOT00000088624
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_+_172273459 | 2.20 |
ENSRNOT00000017957
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr3_+_20641664 | 2.09 |
ENSRNOT00000044699
|
AABR07051741.1
|
|
| chr18_-_19275273 | 2.08 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr17_+_56935451 | 2.06 |
ENSRNOT00000058966
|
RGD1564129
|
similar to hypothetical protein 4930474N05 |
| chr9_+_72052966 | 1.76 |
ENSRNOT00000021099
|
Pth2r
|
parathyroid hormone 2 receptor |
| chr2_-_206699105 | 1.68 |
ENSRNOT00000088095
ENSRNOT00000026952 |
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr2_-_192671059 | 1.61 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr2_+_12102487 | 1.61 |
ENSRNOT00000089209
ENSRNOT00000072155 |
LOC100912538
|
centrin-3-like |
| chr3_+_8450275 | 1.57 |
ENSRNOT00000020073
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr2_-_196527127 | 1.57 |
ENSRNOT00000028709
|
Setdb1
|
SET domain, bifurcated 1 |
| chr10_-_13580821 | 1.52 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr3_+_8450612 | 1.41 |
ENSRNOT00000040457
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr5_+_135962911 | 1.34 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr2_+_225645568 | 1.28 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr1_-_197886759 | 1.27 |
ENSRNOT00000074274
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr2_+_9683570 | 1.19 |
ENSRNOT00000074157
|
Cetn3
|
centrin 3 |
| chr5_-_6186329 | 1.11 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr4_+_61924013 | 1.07 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr4_-_170932618 | 1.04 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr20_-_10968432 | 0.99 |
ENSRNOT00000001593
|
Cstb
|
cystatin B |
| chr6_+_96871625 | 0.92 |
ENSRNOT00000012361
|
Snapc1
|
small nuclear RNA activating complex, polypeptide 1 |
| chr7_-_2712723 | 0.91 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr2_+_27107318 | 0.91 |
ENSRNOT00000019553
|
Arhgef26
|
Rho guanine nucleotide exchange factor 26 |
| chr9_+_17122284 | 0.75 |
ENSRNOT00000077749
|
Polr1c
|
RNA polymerase I subunit C |
| chr9_-_10734073 | 0.66 |
ENSRNOT00000071199
|
Kdm4b
|
lysine demethylase 4B |
| chr10_+_4945911 | 0.46 |
ENSRNOT00000003420
|
Prm1
|
protamine 1 |
| chr2_-_196526886 | 0.45 |
ENSRNOT00000077325
|
Setdb1
|
SET domain, bifurcated 1 |
| chr2_+_198040536 | 0.33 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr3_-_20479999 | 0.30 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr10_+_86860685 | 0.26 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr7_-_59587382 | 0.20 |
ENSRNOT00000091427
ENSRNOT00000066396 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 5.0 | 14.9 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 4.9 | 19.7 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 4.4 | 13.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 4.2 | 16.8 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 3.5 | 17.7 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 2.8 | 39.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.6 | 7.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.6 | 10.6 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 2.5 | 17.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 2.5 | 20.3 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 2.2 | 13.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 2.2 | 17.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 2.1 | 25.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 2.1 | 6.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 2.0 | 6.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.0 | 11.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.8 | 5.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.7 | 10.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.6 | 10.9 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 1.5 | 10.2 | GO:0050882 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Wnt signaling pathway, calcium modulating pathway(GO:0007223) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) voluntary musculoskeletal movement(GO:0050882) |
| 1.5 | 59.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 1.2 | 4.8 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.1 | 4.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.9 | 2.8 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.9 | 2.7 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.8 | 3.3 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.7 | 9.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.7 | 23.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.7 | 3.5 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.7 | 3.3 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.6 | 3.6 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.6 | 8.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.6 | 4.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.6 | 3.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.5 | 2.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 4.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.4 | 1.3 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.4 | 9.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.4 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.4 | 20.3 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.4 | 4.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 11.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.3 | 6.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 3.0 | GO:0090385 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 0.9 | GO:2000318 | positive regulation of memory T cell differentiation(GO:0043382) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.3 | 4.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.3 | 5.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 5.2 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.3 | 19.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.3 | 22.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 4.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 5.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 1.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 4.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 2.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 2.3 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 1.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 8.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 11.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 4.2 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.0 | 3.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.7 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 2.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 10.9 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 2.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 4.4 | 13.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.8 | 19.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 2.5 | 10.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 2.5 | 14.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 2.2 | 13.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.0 | 85.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.7 | 39.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.2 | 10.6 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.0 | 4.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.7 | 17.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 17.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 2.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 4.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 6.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 3.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 5.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 3.0 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 4.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 15.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 5.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 17.9 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 4.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 33.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 8.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 11.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 5.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 15.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 11.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 6.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 6.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 7.9 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 3.6 | 10.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 3.3 | 13.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.5 | 10.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.4 | 16.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 2.3 | 20.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 2.2 | 13.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 2.1 | 6.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 2.0 | 15.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 2.0 | 23.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.8 | 17.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.8 | 8.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.7 | 11.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.5 | 10.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.3 | 17.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.2 | 14.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.1 | 8.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.0 | 20.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.9 | 19.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.8 | 13.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 4.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.7 | 7.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 6.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.6 | 4.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.6 | 2.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.5 | 10.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.5 | 59.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.5 | 2.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 25.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 10.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 6.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 3.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 21.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 19.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 4.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 9.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 2.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 5.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 1.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 7.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 2.2 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 3.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 8.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 2.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 5.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 3.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 5.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 4.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 17.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.2 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 14.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 11.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 10.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 5.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 2.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 20.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 1.0 | 19.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 10.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 10.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 7.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 20.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 8.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 37.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 9.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 3.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 39.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.8 | 20.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.7 | 8.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.7 | 10.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 19.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 13.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 10.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 19.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 16.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 17.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 4.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 9.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 17.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 3.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 11.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 9.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 10.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |