Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
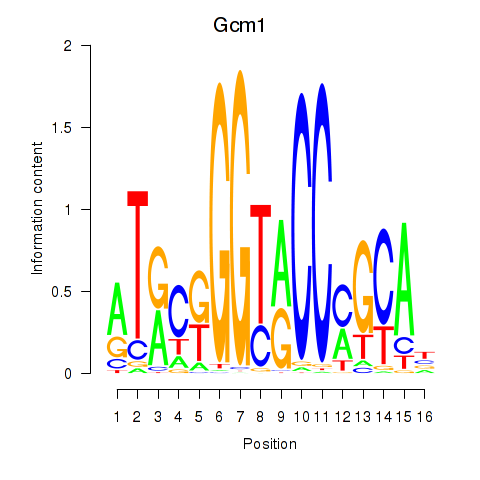
Results for Gcm1
Z-value: 0.82
Transcription factors associated with Gcm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm1
|
ENSRNOG00000007932 | glial cells missing homolog 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gcm1 | rn6_v1_chr8_+_85355766_85355766 | -0.17 | 2.7e-03 | Click! |
Activity profile of Gcm1 motif
Sorted Z-values of Gcm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_16537433 | 28.14 |
ENSRNOT00000048523
|
AABR07051533.2
|
|
| chr11_+_85508300 | 26.59 |
ENSRNOT00000038646
|
AABR07034730.3
|
|
| chr4_-_103369224 | 26.24 |
ENSRNOT00000075709
|
AABR07061057.1
|
|
| chr11_+_85532526 | 26.01 |
ENSRNOT00000036565
|
AABR07034730.2
|
|
| chr17_-_31813716 | 24.70 |
ENSRNOT00000074147
|
AABR07027451.1
|
|
| chr11_+_85561460 | 23.13 |
ENSRNOT00000075455
|
AABR07072262.1
|
|
| chr11_+_85618714 | 22.95 |
ENSRNOT00000074614
|
AC109901.1
|
|
| chr3_+_19141133 | 20.40 |
ENSRNOT00000058323
|
AABR07051675.1
|
|
| chr14_-_78377825 | 19.26 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr3_+_19128400 | 16.35 |
ENSRNOT00000074272
|
AABR07051673.1
|
|
| chr14_-_20953095 | 15.57 |
ENSRNOT00000004440
|
Dck
|
deoxycytidine kinase |
| chr14_-_3389943 | 14.71 |
ENSRNOT00000037101
|
Ephx4
|
epoxide hydrolase 4 |
| chr5_+_154522119 | 12.66 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr11_+_85633243 | 12.08 |
ENSRNOT00000045807
|
LOC682352
|
Ig lambda chain V-VI region AR-like |
| chr16_+_81616604 | 11.77 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr1_+_221773254 | 11.16 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr3_+_19174027 | 10.64 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr3_+_33440191 | 9.81 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr5_-_75676584 | 9.51 |
ENSRNOT00000044348
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr7_-_115910522 | 9.39 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr7_-_12346475 | 9.12 |
ENSRNOT00000060708
|
Mum1
|
melanoma associated antigen (mutated) 1 |
| chr10_+_105861743 | 8.76 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr1_-_259674425 | 8.53 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chr1_-_259089632 | 8.43 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr1_+_53360157 | 7.30 |
ENSRNOT00000017809
|
Rps6ka2
|
ribosomal protein S6 kinase A2 |
| chr15_+_34187223 | 7.29 |
ENSRNOT00000024978
|
Cpne6
|
copine 6 |
| chr8_+_7128656 | 7.16 |
ENSRNOT00000038313
|
Pgr
|
progesterone receptor |
| chr16_+_81089292 | 7.07 |
ENSRNOT00000087192
ENSRNOT00000026150 |
Tfdp1
|
transcription factor Dp-1 |
| chr11_-_86276430 | 7.00 |
ENSRNOT00000075164
|
Hira
|
histone cell cycle regulator |
| chr7_-_122247139 | 6.95 |
ENSRNOT00000064515
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr8_+_96551245 | 6.94 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr20_+_50394650 | 5.84 |
ENSRNOT00000076010
ENSRNOT00000077065 ENSRNOT00000073259 |
Popdc3
|
popeye domain containing 3 |
| chr1_+_137799185 | 5.64 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr2_-_166255302 | 5.12 |
ENSRNOT00000016012
|
B3galnt1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr5_-_173233188 | 5.05 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr9_+_47386626 | 4.95 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr7_-_126913585 | 4.58 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr5_+_157165341 | 4.54 |
ENSRNOT00000087072
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr4_-_163649637 | 4.52 |
ENSRNOT00000080873
|
LOC690020
|
similar to killer cell lectin-like receptor, subfamily A, member 17 |
| chrX_+_62363757 | 4.50 |
ENSRNOT00000091240
|
Arx
|
aristaless related homeobox |
| chr1_-_101131413 | 4.42 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr14_-_104523297 | 4.39 |
ENSRNOT00000039523
|
Cep68
|
centrosomal protein 68 |
| chr10_-_11878792 | 3.82 |
ENSRNOT00000064964
|
Cluap1
|
clusterin associated protein 1 |
| chr7_-_12424367 | 3.53 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr8_-_55696601 | 3.44 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr5_+_139468546 | 3.43 |
ENSRNOT00000013193
|
Slfnl1
|
schlafen-like 1 |
| chr1_-_101131012 | 3.38 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr16_+_8823872 | 3.14 |
ENSRNOT00000027186
|
Drgx
|
dorsal root ganglia homeobox |
| chr11_+_86276616 | 3.11 |
ENSRNOT00000071159
|
Mrpl40
|
mitochondrial ribosomal protein L40 |
| chr4_+_38240728 | 3.09 |
ENSRNOT00000047029
|
Phf14
|
PHD finger protein 14 |
| chr8_+_119121669 | 2.91 |
ENSRNOT00000028479
|
Prss46
|
protease, serine, 46 |
| chrX_+_18998532 | 2.86 |
ENSRNOT00000004213
|
AABR07037343.1
|
|
| chr16_+_34795971 | 2.66 |
ENSRNOT00000043510
|
LOC100912321
|
myeloid-associated differentiation marker-like |
| chr9_+_118586179 | 2.62 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chrX_+_62363953 | 2.53 |
ENSRNOT00000083362
|
Arx
|
aristaless related homeobox |
| chr1_+_220423426 | 2.51 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr3_-_122947075 | 2.51 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr6_-_137084739 | 2.34 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr3_-_8924032 | 2.22 |
ENSRNOT00000023527
ENSRNOT00000085042 |
Sh3glb2
|
SH3 domain-containing GRB2-like endophilin B2 |
| chr13_-_111849653 | 2.21 |
ENSRNOT00000006395
|
Diexf
|
digestive organ expansion factor homolog (zebrafish) |
| chr4_-_113764532 | 2.18 |
ENSRNOT00000009269
|
Sema4f
|
ssemaphorin 4F |
| chr6_+_135610743 | 2.17 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr8_-_48736506 | 2.16 |
ENSRNOT00000016227
|
Ccdc84
|
coiled-coil domain containing 84 |
| chr3_+_108944141 | 2.06 |
ENSRNOT00000034950
|
Fam98b
|
family with sequence similarity 98, member B |
| chr2_+_204886202 | 2.00 |
ENSRNOT00000022200
|
Ngf
|
nerve growth factor |
| chr10_+_61685645 | 1.79 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr19_-_17346808 | 1.76 |
ENSRNOT00000017361
|
Rbl2
|
RB transcriptional corepressor like 2 |
| chr2_+_266141581 | 1.65 |
ENSRNOT00000078187
ENSRNOT00000051951 |
Rpe65
|
RPE65, retinoid isomerohydrolase |
| chr2_+_57206613 | 1.62 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr19_+_37127508 | 1.51 |
ENSRNOT00000019656
|
Cbfb
|
core-binding factor, beta subunit |
| chr5_-_153252021 | 1.40 |
ENSRNOT00000023221
|
Tmem50a
|
transmembrane protein 50A |
| chr2_-_157066781 | 1.30 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr15_+_36897999 | 1.15 |
ENSRNOT00000076030
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr14_+_104191517 | 1.12 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_60988124 | 1.05 |
ENSRNOT00000038294
|
LOC100911247
|
transmembrane protein 186-like |
| chr7_-_14217778 | 1.04 |
ENSRNOT00000038961
|
Ephx3
|
epoxide hydrolase 3 |
| chr9_+_49479023 | 1.04 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr4_-_38240848 | 0.92 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr3_-_95954983 | 0.88 |
ENSRNOT00000006385
|
AABR07053185.1
|
|
| chr4_+_1459363 | 0.70 |
ENSRNOT00000082919
|
Olr1234
|
olfactory receptor 1234 |
| chr11_-_87158597 | 0.70 |
ENSRNOT00000002517
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr6_-_147172022 | 0.58 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr3_-_21061070 | 0.45 |
ENSRNOT00000044461
|
Olr422
|
olfactory receptor 422 |
| chrX_-_25628272 | 0.39 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr15_+_33885106 | 0.29 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr20_+_1110773 | 0.20 |
ENSRNOT00000041697
|
Olr1702
|
olfactory receptor 1702 |
| chr11_+_88376256 | 0.19 |
ENSRNOT00000081776
|
Vpreb1
|
pre-B lymphocyte 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gcm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 2.9 | 11.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.6 | 15.6 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 2.4 | 7.2 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 2.1 | 12.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.9 | 7.8 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 1.9 | 9.5 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 1.7 | 17.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 1.5 | 4.6 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.3 | 3.8 | GO:0021508 | floor plate formation(GO:0021508) |
| 1.2 | 9.4 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 1.2 | 7.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.0 | 7.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 1.0 | 7.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.9 | 5.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.9 | 7.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.7 | 3.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 3.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.5 | 1.6 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.4 | 5.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 4.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 5.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 2.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 2.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 1.0 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.3 | 18.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.3 | 2.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 3.1 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.2 | 7.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.2 | 2.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 4.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 8.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 2.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 1.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 2.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.2 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 8.7 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 2.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.9 | GO:0042100 | B cell proliferation(GO:0042100) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 9.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 3.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 3.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 2.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 7.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 4.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 2.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 2.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 26.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 11.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 21.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 8.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 4.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 5.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.6 | GO:0019136 | pyrimidine nucleoside binding(GO:0001884) deoxynucleoside kinase activity(GO:0019136) |
| 2.8 | 17.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 2.4 | 9.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 2.2 | 8.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.8 | 7.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.0 | 5.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 4.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 7.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.5 | 2.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.5 | 5.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 7.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.5 | 11.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.4 | 6.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.3 | 16.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 2.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 5.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.6 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.2 | 1.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 5.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 7.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 7.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 7.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 11.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 8.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 5.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 11.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 4.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 7.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 9.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 11.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 2.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 7.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 9.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 7.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 12.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 11.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 7.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.5 | 8.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 7.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 2.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 7.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 9.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |