Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
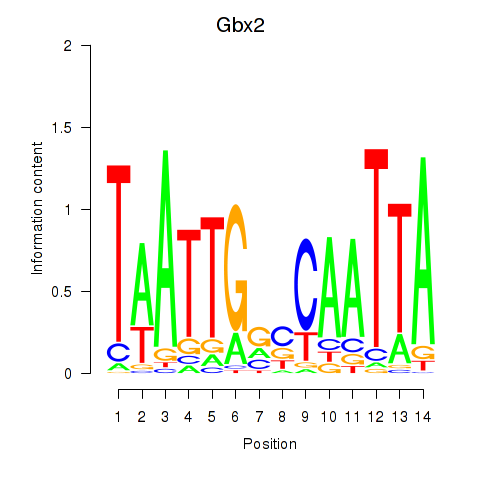
Results for Gbx2
Z-value: 0.68
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSRNOG00000019495 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | rn6_v1_chr9_-_97065817_97065817 | -0.11 | 4.3e-02 | Click! |
Activity profile of Gbx2 motif
Sorted Z-values of Gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_127632265 | 52.01 |
ENSRNOT00000084157
|
Serpina1
|
serpin family A member 1 |
| chr3_+_159368273 | 51.40 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_+_147713892 | 47.83 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr1_+_250426158 | 29.25 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr1_+_248428099 | 27.02 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr4_-_169036950 | 20.54 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr4_-_23718047 | 17.89 |
ENSRNOT00000011359
|
RGD1564345
|
similar to RIKEN cDNA 4921511H03 |
| chr14_-_21748356 | 17.54 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr10_+_103206014 | 16.99 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr1_-_174047572 | 14.20 |
ENSRNOT00000019597
|
Stk33
|
serine/threonine kinase 33 |
| chrY_-_1398030 | 13.32 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr12_+_47179664 | 11.74 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr2_+_54466280 | 11.00 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr5_+_78334284 | 10.78 |
ENSRNOT00000020366
|
Bspry
|
B-box and SPRY domain containing |
| chr5_+_28480023 | 10.48 |
ENSRNOT00000075067
ENSRNOT00000093754 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr7_-_107223047 | 10.17 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chrX_-_14783792 | 8.53 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr2_-_173563273 | 8.26 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr8_+_119083900 | 7.05 |
ENSRNOT00000051947
ENSRNOT00000048655 |
Prss44
|
protease, serine, 44 |
| chr2_+_220432037 | 6.72 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr2_+_72006099 | 6.66 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr14_-_43143973 | 6.11 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr4_-_11497531 | 5.83 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_+_48106099 | 5.81 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chrX_+_84064427 | 5.51 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr10_+_11912543 | 5.44 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr8_-_45137893 | 5.16 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr8_+_37557067 | 4.97 |
ENSRNOT00000073109
|
LOC688335
|
similar to Spleen protein 1 precursor (RSP-1) |
| chr2_-_122641409 | 4.76 |
ENSRNOT00000068334
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr2_+_208738132 | 4.73 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr8_+_38396532 | 4.60 |
ENSRNOT00000071278
|
LOC100360143
|
rCG64164-like |
| chr8_+_38685415 | 4.60 |
ENSRNOT00000073660
|
LOC688335
|
similar to Spleen protein 1 precursor (RSP-1) |
| chr13_-_83457888 | 4.58 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr2_-_198706428 | 4.50 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr17_+_25082056 | 4.35 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr1_+_220071811 | 4.28 |
ENSRNOT00000090642
|
AC126581.1
|
|
| chr12_-_46493203 | 4.07 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr17_-_62299655 | 3.41 |
ENSRNOT00000024855
|
Gjd4
|
gap junction protein, delta 4 |
| chr16_+_90613870 | 3.34 |
ENSRNOT00000079334
|
Shcbp1
|
SHC binding and spindle associated 1 |
| chr4_-_27726943 | 3.25 |
ENSRNOT00000068499
|
LOC500007
|
similar to EF hand calcium binding domain 1 |
| chr5_-_40867023 | 3.24 |
ENSRNOT00000011576
|
Manea
|
mannosidase, endo-alpha |
| chr1_-_78180216 | 3.24 |
ENSRNOT00000071576
|
C5ar2
|
complement component 5a receptor 2 |
| chr3_+_147952879 | 3.09 |
ENSRNOT00000031922
|
Defb20
|
defensin beta 20 |
| chr1_-_214414763 | 2.86 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr14_+_22597103 | 2.60 |
ENSRNOT00000048482
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr6_-_108660063 | 2.60 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr8_-_41445722 | 2.57 |
ENSRNOT00000011381
|
LOC171573
|
spleen protein 1 precursor |
| chr8_+_117117430 | 2.05 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_+_79638696 | 1.92 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr4_+_143911791 | 1.90 |
ENSRNOT00000081511
|
AABR07061780.1
|
|
| chr2_-_189333015 | 1.82 |
ENSRNOT00000076424
ENSRNOT00000076952 |
Hax1
|
HCLS1 associated protein X-1 |
| chr2_-_189333322 | 1.80 |
ENSRNOT00000073599
ENSRNOT00000071253 ENSRNOT00000076325 |
Hax1
|
HCLS1 associated protein X-1 |
| chr14_-_106248539 | 1.57 |
ENSRNOT00000067668
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chrX_-_124252447 | 1.52 |
ENSRNOT00000061546
|
Rhox12
|
reproductive homeobox 12 |
| chr3_+_138508899 | 1.44 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr8_+_38240206 | 1.22 |
ENSRNOT00000030635
|
LOC100362690
|
rCG64164-like |
| chr5_+_135574172 | 1.16 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr18_+_16146447 | 0.94 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr18_+_51523758 | 0.83 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr20_+_20519031 | 0.72 |
ENSRNOT00000000780
|
AABR07044783.1
|
|
| chr9_+_73378057 | 0.39 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr8_+_69484174 | 0.39 |
ENSRNOT00000077290
ENSRNOT00000056518 |
RGD1562747
|
similar to RIKEN cDNA 1110012L19 |
| chr3_-_138683318 | 0.22 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr12_-_44520341 | 0.21 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
| chr5_-_9035811 | 0.03 |
ENSRNOT00000082023
|
Mcmdc2
|
minichromosome maintenance domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 52.0 | GO:0033986 | response to methanol(GO:0033986) |
| 9.6 | 47.8 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 9.5 | 38.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 7.3 | 29.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 1.1 | 10.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.1 | 3.2 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.0 | 11.7 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.7 | 5.8 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.7 | 2.0 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.7 | 3.4 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.7 | 6.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.7 | 8.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.6 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.4 | 51.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.3 | 5.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 3.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 4.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 2.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 2.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.9 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 3.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 4.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 17.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 14.2 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 13.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 4.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 6.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 18.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 3.3 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 10.8 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 29.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 1.6 | 11.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 5.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.6 | 5.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 2.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.4 | 27.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 17.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 4.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 6.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 17.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 66.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 3.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 5.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 11.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 43.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 10.8 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 10.2 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 3.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 38.9 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 27.0 | GO:0005534 | galactose binding(GO:0005534) |
| 2.7 | 51.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.7 | 47.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.7 | 6.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.5 | 6.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.5 | 5.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.1 | 3.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.9 | 11.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.9 | 10.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.6 | 2.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 52.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 8.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 20.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.4 | 1.6 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.4 | 29.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 3.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 4.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 5.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 17.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 3.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 13.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 2.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 27.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 7.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 44.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 6.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 21.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.1 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 27.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 11.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 52.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 6.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 4.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 4.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 29.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 3.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |