Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
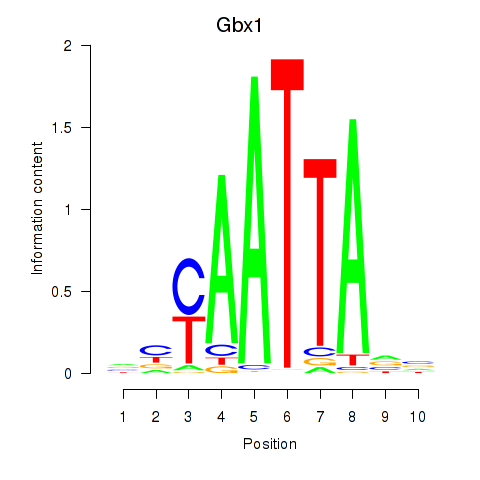
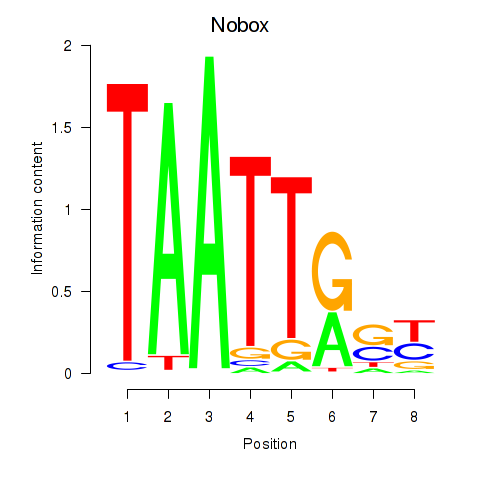
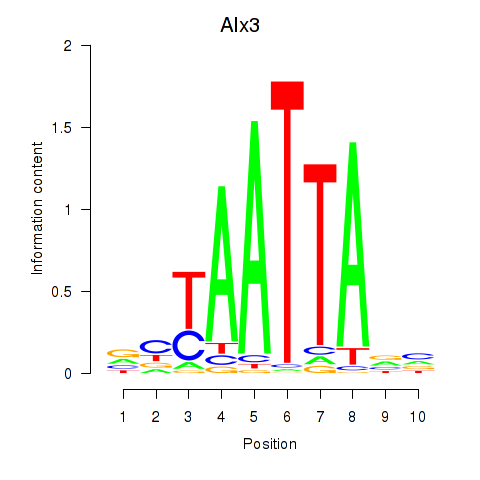
Results for Gbx1_Nobox_Alx3
Z-value: 0.50
Transcription factors associated with Gbx1_Nobox_Alx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx1
|
ENSRNOG00000013369 | gastrulation brain homeobox 1 |
|
Nobox
|
ENSRNOG00000025720 | NOBOX oogenesis homeobox |
|
Alx3
|
ENSRNOG00000018290 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nobox | rn6_v1_chr4_-_72756887_72756887 | 0.10 | 6.6e-02 | Click! |
| Gbx1 | rn6_v1_chr4_+_7173961_7173961 | -0.05 | 3.3e-01 | Click! |
| Alx3 | rn6_v1_chr2_+_210381829_210381829 | -0.05 | 3.6e-01 | Click! |
Activity profile of Gbx1_Nobox_Alx3 motif
Sorted Z-values of Gbx1_Nobox_Alx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_7058314 | 17.44 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr1_+_279633671 | 16.18 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr2_+_54466280 | 15.82 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_-_157304653 | 14.02 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr13_-_76049363 | 13.90 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr16_+_10267482 | 13.84 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr9_+_20241062 | 13.21 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr2_-_219262901 | 12.40 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr8_+_106449321 | 11.86 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr6_+_2216623 | 10.89 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr10_-_93675991 | 9.40 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr9_-_81566096 | 9.39 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr2_-_60657712 | 9.18 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr4_-_17594598 | 9.12 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr10_-_87248572 | 8.83 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr4_+_22898527 | 8.45 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr9_+_53013413 | 7.81 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr16_+_2634603 | 7.76 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr5_+_61474000 | 7.16 |
ENSRNOT00000013930
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr14_+_76732650 | 7.09 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr10_-_87232723 | 6.84 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr2_-_250862419 | 6.76 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr5_+_117698764 | 6.75 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr4_+_6827429 | 6.49 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr20_+_44680449 | 6.23 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr3_-_165700489 | 6.04 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr18_-_16543992 | 5.96 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr19_+_25043680 | 5.91 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr7_-_68549763 | 5.90 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chrX_+_84064427 | 5.85 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr7_-_68512397 | 5.51 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr4_+_147832136 | 5.49 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr1_+_140998240 | 5.44 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr11_-_71284939 | 5.26 |
ENSRNOT00000002421
|
AABR07034428.1
|
|
| chr2_-_138833933 | 5.04 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr5_-_115387377 | 5.01 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr3_-_57607683 | 4.98 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr8_+_85503224 | 4.91 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr14_+_59611434 | 4.86 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr9_-_81566642 | 4.84 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr7_-_143228060 | 4.78 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr8_-_53816447 | 4.77 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr4_-_100252755 | 4.76 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr14_-_16903242 | 4.72 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr2_+_80269661 | 4.66 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr16_+_50049828 | 4.64 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr5_+_50381244 | 4.59 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr8_+_47674321 | 4.57 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr17_+_72160735 | 4.56 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr5_-_78985990 | 4.51 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr2_+_72006099 | 4.51 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr11_-_14304603 | 4.42 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr10_-_87468711 | 4.33 |
ENSRNOT00000039983
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr1_-_43638161 | 4.32 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_81694808 | 4.24 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr3_-_90751055 | 4.24 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr4_-_88684415 | 4.23 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr1_+_185210922 | 4.21 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr20_-_30947484 | 4.17 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr11_+_45751812 | 4.06 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr6_-_78549669 | 4.05 |
ENSRNOT00000009940
|
Foxa1
|
forkhead box A1 |
| chr1_-_89539210 | 4.04 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr3_-_66417741 | 4.01 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr6_-_108660063 | 3.98 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr8_+_57983556 | 3.98 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr4_-_80395502 | 3.97 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr11_+_61609370 | 3.83 |
ENSRNOT00000088880
ENSRNOT00000082533 |
Gramd1c
|
GRAM domain containing 1C |
| chr4_-_21920651 | 3.79 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr10_-_87529599 | 3.75 |
ENSRNOT00000074099
|
Krtap2-1
|
keratin associated protein 2-1 |
| chr14_-_19072677 | 3.63 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr2_+_252090669 | 3.58 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr7_+_60316290 | 3.57 |
ENSRNOT00000085673
|
Lyc2
|
lysozyme C type 2 |
| chr8_+_41350182 | 3.56 |
ENSRNOT00000072823
|
LOC100911758
|
olfactory receptor 143-like |
| chr6_-_115616766 | 3.43 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr9_-_4876023 | 3.36 |
ENSRNOT00000065839
|
RGD1559960
|
similar to Sulfotransferase K1 (rSULT1C2) |
| chr6_-_114476723 | 3.35 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr10_-_87286387 | 3.31 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr7_-_145062956 | 3.23 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr10_+_62981297 | 3.23 |
ENSRNOT00000031618
|
Efcab5
|
EF-hand calcium binding domain 5 |
| chr14_-_21252538 | 3.17 |
ENSRNOT00000005003
|
Ambn
|
ameloblastin |
| chr16_-_73152921 | 3.16 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr3_-_103745236 | 3.12 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr10_+_82032656 | 3.12 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr7_+_60316045 | 3.08 |
ENSRNOT00000064594
|
Lyc2
|
lysozyme C type 2 |
| chr7_-_143497108 | 3.06 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr13_+_79378733 | 3.00 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr8_-_114617466 | 2.96 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr6_-_77421286 | 2.95 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr17_+_25082056 | 2.90 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr1_-_227932603 | 2.88 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr6_+_132510757 | 2.87 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr12_-_45801842 | 2.87 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chrX_+_105011489 | 2.87 |
ENSRNOT00000085068
|
Arl13a
|
ADP ribosylation factor like GTPase 13A |
| chr10_-_88670430 | 2.78 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr13_-_90405591 | 2.77 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr17_+_47301511 | 2.75 |
ENSRNOT00000092051
ENSRNOT00000087178 |
Nme8
|
NME/NM23 family member 8 |
| chr12_+_18679789 | 2.69 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr10_+_87782376 | 2.68 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chr17_+_47302272 | 2.67 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr14_+_96499520 | 2.65 |
ENSRNOT00000074692
|
AABR07016310.1
|
|
| chr8_-_12993651 | 2.64 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr3_+_113319456 | 2.60 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr6_-_36876805 | 2.58 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr20_-_10013190 | 2.58 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr13_+_82438697 | 2.51 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr11_-_46693565 | 2.50 |
ENSRNOT00000044900
|
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr19_-_58735173 | 2.48 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr13_+_91054974 | 2.47 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr9_+_81566074 | 2.40 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr1_+_225037737 | 2.33 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr10_+_91217079 | 2.29 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr13_-_95250235 | 2.23 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chrX_+_118197217 | 2.23 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr19_-_25961666 | 2.21 |
ENSRNOT00000004091
|
Calr
|
calreticulin |
| chr8_-_40137390 | 2.13 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr16_-_25192675 | 2.13 |
ENSRNOT00000032289
|
March1
|
membrane associated ring-CH-type finger 1 |
| chrX_+_74200972 | 2.09 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_+_87066289 | 2.07 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr5_-_12526962 | 2.04 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr14_-_104612597 | 2.04 |
ENSRNOT00000007002
|
Slc1a4
|
solute carrier family 1 member 4 |
| chr1_-_199159125 | 2.03 |
ENSRNOT00000025618
|
Bcl7c
|
BCL tumor suppressor 7C |
| chr14_+_91588940 | 2.03 |
ENSRNOT00000057067
|
RGD1309870
|
hypothetical LOC289778 |
| chrX_-_16532089 | 2.00 |
ENSRNOT00000093156
|
Dgkk
|
diacylglycerol kinase kappa |
| chr3_+_15560712 | 1.99 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_-_73130740 | 1.98 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr2_-_122641409 | 1.96 |
ENSRNOT00000068334
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr3_+_19174027 | 1.95 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chrX_+_74205842 | 1.89 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr10_+_70242874 | 1.85 |
ENSRNOT00000010924
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr5_+_133819726 | 1.81 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr1_-_166912524 | 1.81 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr3_-_76696107 | 1.77 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr3_-_45169118 | 1.75 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr5_+_117583502 | 1.73 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
| chrX_+_14578264 | 1.72 |
ENSRNOT00000038994
|
Cybb
|
cytochrome b-245 beta chain |
| chr1_-_24604400 | 1.72 |
ENSRNOT00000081175
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chrX_+_148421627 | 1.68 |
ENSRNOT00000016754
|
Ctag2
|
cancer/testis antigen 2 |
| chrX_-_124464963 | 1.67 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr1_+_278557792 | 1.66 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr7_+_44009069 | 1.64 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chrY_-_1398030 | 1.62 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr7_-_28711761 | 1.61 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr6_-_128149220 | 1.59 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr20_+_3558827 | 1.59 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_16846412 | 1.54 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chr17_-_45746753 | 1.53 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr5_+_133620706 | 1.53 |
ENSRNOT00000053202
|
Gm12830
|
predicted gene 12830 |
| chr2_+_154604832 | 1.51 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr11_-_782954 | 1.50 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr4_+_2055615 | 1.49 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr4_+_148476943 | 1.48 |
ENSRNOT00000017689
|
Olr823
|
olfactory receptor 823 |
| chr3_+_19045214 | 1.48 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr4_-_30380119 | 1.47 |
ENSRNOT00000036460
|
Pon2
|
paraoxonase 2 |
| chr12_-_17186679 | 1.47 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr3_+_94530586 | 1.46 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr4_-_50312608 | 1.45 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr4_+_117962319 | 1.43 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr1_-_89269930 | 1.43 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr5_-_28130803 | 1.42 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr3_+_149643481 | 1.42 |
ENSRNOT00000020713
|
Bpifa5
|
BPI fold containing family A, member 5 |
| chr1_-_101095594 | 1.38 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr13_+_56546021 | 1.38 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr6_+_8886591 | 1.36 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr12_+_22727335 | 1.36 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr10_-_86004096 | 1.35 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr11_+_31539016 | 1.30 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr2_-_35104963 | 1.28 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr6_-_111417813 | 1.27 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr4_-_148845267 | 1.24 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr12_-_44911147 | 1.22 |
ENSRNOT00000071074
ENSRNOT00000046190 |
Ksr2
|
kinase suppressor of ras 2 |
| chr5_-_93244202 | 1.21 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr7_-_143167772 | 1.21 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr8_+_40410604 | 1.21 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr1_-_173764246 | 1.18 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr9_+_4107246 | 1.18 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr13_+_49005405 | 1.17 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr19_-_44095250 | 1.15 |
ENSRNOT00000074824
|
Tmem170a
|
transmembrane protein 170A |
| chr11_-_54567357 | 1.14 |
ENSRNOT00000046890
|
Retnlb
|
resistin like beta |
| chr3_+_55461420 | 1.13 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chrY_-_1305026 | 1.13 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_-_28131133 | 1.13 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr5_+_135574172 | 1.13 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr12_-_22726982 | 1.12 |
ENSRNOT00000001921
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr14_-_21128505 | 1.12 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chrX_-_35777243 | 1.12 |
ENSRNOT00000040912
|
Rs1
|
retinoschisin 1 |
| chr5_-_12172009 | 1.09 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chrX_-_18341467 | 1.07 |
ENSRNOT00000045802
|
Spin2a
|
spindlin family, member 2A |
| chr6_-_72805848 | 1.05 |
ENSRNOT00000009040
|
Gpr33
|
G protein-coupled receptor 33 |
| chr9_+_117795132 | 1.04 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr1_+_169966190 | 1.00 |
ENSRNOT00000023307
|
Olr190
|
olfactory receptor 190 |
| chr16_-_14348046 | 1.00 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr10_+_56601288 | 1.00 |
ENSRNOT00000023724
|
Gabarap
|
GABA type A receptor-associated protein |
| chr7_+_41475163 | 0.99 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr7_+_58570579 | 0.97 |
ENSRNOT00000081227
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr3_-_122947075 | 0.97 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr15_+_44441856 | 0.92 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr1_-_167971151 | 0.89 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr10_-_13446135 | 0.89 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr3_-_10196626 | 0.88 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr12_-_22138382 | 0.88 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx1_Nobox_Alx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 4.4 | 17.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 2.5 | 12.4 | GO:0061743 | motor learning(GO:0061743) |
| 2.3 | 11.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.6 | 4.8 | GO:1903595 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.5 | 11.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.4 | 4.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.4 | 6.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.3 | 4.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 1.2 | 4.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 1.2 | 8.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.0 | 4.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.0 | 10.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 1.0 | 3.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.9 | 2.8 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.9 | 2.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.8 | 3.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.8 | 6.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.8 | 4.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) alveolar secondary septum development(GO:0061144) |
| 0.7 | 2.2 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.7 | 2.2 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.7 | 13.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.7 | 2.7 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.6 | 6.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.6 | 1.7 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.5 | 2.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.5 | 1.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 7.9 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.5 | 5.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.5 | 2.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 2.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 1.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 2.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 2.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.4 | 1.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 1.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.4 | GO:0002877 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.4 | 8.4 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.3 | 2.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.3 | 6.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.3 | 1.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 0.9 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.3 | 4.9 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.3 | 3.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 0.8 | GO:0060166 | olfactory pit development(GO:0060166) Harderian gland development(GO:0070384) |
| 0.3 | 6.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 9.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.2 | 3.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 1.4 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.2 | 6.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.7 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 5.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 2.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.2 | 3.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 1.8 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.6 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 2.6 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.2 | 13.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 14.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 1.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 4.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 2.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.9 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.6 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.2 | 0.5 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.1 | 1.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.5 | GO:0072592 | positive regulation of integrin biosynthetic process(GO:0045726) pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) oxygen metabolic process(GO:0072592) |
| 0.1 | 3.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 2.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 4.4 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 2.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.5 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.7 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 1.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.6 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 1.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 2.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 7.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.5 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.9 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 1.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.9 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.7 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.3 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 5.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.8 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 2.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 2.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 2.3 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 3.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 2.5 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 4.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 3.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.0 | 1.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 3.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 15.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.6 | 4.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.1 | 3.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.9 | 2.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.9 | 5.4 | GO:0036157 | outer dynein arm(GO:0036157) sperm cytoplasmic droplet(GO:0097598) |
| 0.8 | 2.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.8 | 5.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.6 | 2.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 2.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 14.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 6.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 6.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 4.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 19.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.9 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.2 | 2.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 15.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 4.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 11.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 8.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 4.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.9 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 6.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 10.3 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 7.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.1 | 17.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.9 | 4.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.8 | 3.2 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.8 | 6.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.7 | 9.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 2.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.6 | 17.4 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.6 | 2.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 2.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.5 | 4.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.5 | 6.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 2.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.5 | 1.5 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.5 | 2.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 1.3 | GO:0019961 | interferon binding(GO:0019961) |
| 0.4 | 5.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 4.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.4 | 1.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.4 | 2.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 6.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 4.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.3 | 5.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 2.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 1.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 4.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 5.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 1.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 2.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 13.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 2.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 4.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 1.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.2 | 17.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 4.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 13.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 4.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 2.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 3.0 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 8.7 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 6.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 3.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 4.8 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 2.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 17.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 8.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 6.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 11.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 5.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 1.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 11.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 7.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 6.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 9.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 7.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 11.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 6.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 5.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.5 | 18.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 9.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.5 | 5.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 4.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 6.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 2.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 7.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 4.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 1.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 3.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 4.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 4.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 2.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 6.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 2.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |