Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Gata6
Z-value: 0.44
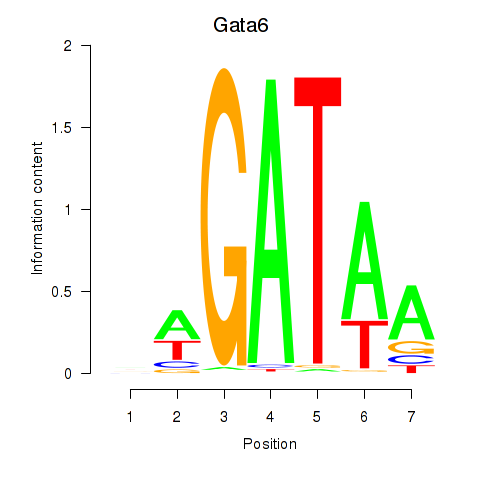
Transcription factors associated with Gata6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata6
|
ENSRNOG00000023433 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata6 | rn6_v1_chr18_+_2416552_2416552 | 0.01 | 8.6e-01 | Click! |
Activity profile of Gata6 motif
Sorted Z-values of Gata6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114423533 | 20.36 |
ENSRNOT00000091221
|
Slc2a2
|
solute carrier family 2 member 2 |
| chr4_+_119225040 | 18.86 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chr8_+_50525091 | 13.62 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr19_+_27404712 | 12.37 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr7_+_11490852 | 12.05 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr2_-_100249811 | 11.47 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr16_+_10267482 | 9.95 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr1_+_279798187 | 9.56 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr3_+_79940561 | 9.52 |
ENSRNOT00000016652
|
Mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_7961207 | 8.54 |
ENSRNOT00000007174
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr16_-_49820235 | 6.69 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_207766250 | 5.36 |
ENSRNOT00000031397
|
Clrn3
|
clarin 3 |
| chr6_+_7961413 | 5.25 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr17_-_84247038 | 3.91 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr1_-_207811008 | 3.12 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr11_-_44049648 | 2.39 |
ENSRNOT00000002257
|
Cpox
|
coproporphyrinogen oxidase |
| chr10_+_70262361 | 2.34 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr4_-_50312608 | 1.99 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr15_+_32817343 | 1.99 |
ENSRNOT00000073853
|
AABR07017902.1
|
|
| chr2_+_42726809 | 1.76 |
ENSRNOT00000036860
|
AABR07008097.1
|
|
| chr1_+_199720038 | 1.67 |
ENSRNOT00000027297
|
Ahsp
|
alpha hemoglobin stabilizing protein |
| chr2_+_241029693 | 1.64 |
ENSRNOT00000033413
|
Slc39a8
|
solute carrier family 39 member 8 |
| chr10_-_31493419 | 1.07 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr1_+_214664537 | 0.98 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr13_-_45040593 | 0.87 |
ENSRNOT00000004908
|
Lct
|
lactase |
| chr2_-_77784522 | 0.77 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr2_-_259382765 | 0.16 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr3_+_76842898 | 0.03 |
ENSRNOT00000085537
|
Olr636
|
olfactory receptor 636 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 31.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 4.5 | 13.6 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 4.1 | 20.4 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 2.3 | 13.8 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.3 | 3.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.0 | 9.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 1.0 | 9.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 12.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.5 | 2.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 9.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 1.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 2.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 6.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.9 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.1 | 1.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 11.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 2.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.7 | GO:0006457 | protein folding(GO:0006457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 2.7 | 13.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.1 | 9.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 20.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 22.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 6.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 9.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 8.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 3.1 | 12.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.7 | 18.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 2.3 | 20.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 9.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.8 | 12.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.7 | 13.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.5 | 9.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 2.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 0.9 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.2 | 1.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 3.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 9.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 11.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 34.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 9.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 14.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 27.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 1.2 | 20.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 11.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 9.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |