Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
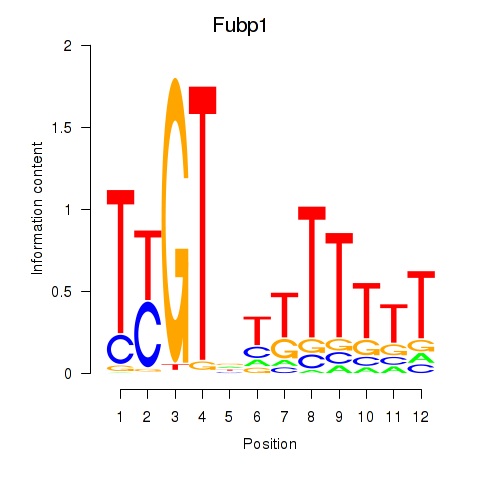
Results for Fubp1
Z-value: 0.54
Transcription factors associated with Fubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fubp1
|
ENSRNOG00000043370 | far upstream element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fubp1 | rn6_v1_chr2_+_257425679_257425679 | 0.24 | 1.2e-05 | Click! |
Activity profile of Fubp1 motif
Sorted Z-values of Fubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32973695 | 18.62 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr2_+_114386019 | 13.69 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr13_-_51784639 | 12.86 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_142689026 | 12.44 |
ENSRNOT00000010007
|
Fhl3
|
four and a half LIM domains 3 |
| chr1_+_68436917 | 10.56 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr4_-_150185044 | 10.40 |
ENSRNOT00000019778
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr6_-_146195819 | 10.24 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr17_+_44766313 | 9.38 |
ENSRNOT00000091688
|
Hist1h1b
|
histone cluster 1 H1 family member b |
| chr1_-_57815038 | 9.11 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chrX_-_29648359 | 8.30 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr7_+_144565392 | 8.12 |
ENSRNOT00000021589
|
Hoxc11
|
homeobox C11 |
| chr2_+_207262934 | 8.06 |
ENSRNOT00000016833
|
Ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr3_-_52447622 | 7.06 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr2_-_211831551 | 6.80 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr3_-_11382004 | 6.49 |
ENSRNOT00000047921
ENSRNOT00000064039 |
Dnm1
|
dynamin 1 |
| chrX_+_105575759 | 6.48 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chrX_+_78042859 | 6.37 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr3_-_61488696 | 5.96 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr20_+_46519431 | 5.45 |
ENSRNOT00000077765
|
AABR07045405.1
|
|
| chrX_-_76925195 | 5.43 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chrX_+_105573909 | 5.00 |
ENSRNOT00000029664
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr9_+_46840992 | 4.89 |
ENSRNOT00000019415
|
Il1r2
|
interleukin 1 receptor type 2 |
| chr16_+_71629525 | 4.62 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr14_-_87701884 | 4.57 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr17_-_11916026 | 4.52 |
ENSRNOT00000046385
|
RGD1561671
|
similar to RIKEN cDNA 2900010M23 |
| chr10_+_15156207 | 4.21 |
ENSRNOT00000020363
|
Ccdc78
|
coiled-coil domain containing 78 |
| chr1_-_216828581 | 4.10 |
ENSRNOT00000066943
ENSRNOT00000088856 |
Tnfrsf26
|
tumor necrosis factor receptor superfamily, member 26 |
| chr4_-_120041238 | 4.06 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr10_-_85825735 | 3.88 |
ENSRNOT00000055379
|
Fbxo47
|
F-box protein 47 |
| chr1_-_167210919 | 3.79 |
ENSRNOT00000027507
|
Chrna10
|
cholinergic receptor nicotinic alpha 10 subunit |
| chr18_-_81807627 | 3.78 |
ENSRNOT00000020376
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr13_+_90116843 | 3.73 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr2_+_235596123 | 3.61 |
ENSRNOT00000074816
|
Col25a1
|
collagen type XXV alpha 1 chain |
| chrX_-_115496045 | 3.59 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr3_+_80556668 | 3.52 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr13_-_72367980 | 3.50 |
ENSRNOT00000003928
|
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr5_+_164923302 | 3.31 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr20_-_32133431 | 3.26 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr12_-_7186873 | 3.23 |
ENSRNOT00000050438
|
LOC103690996
|
uncharacterized LOC103690996 |
| chr3_+_111699021 | 3.22 |
ENSRNOT00000067865
ENSRNOT00000008528 ENSRNOT00000064395 ENSRNOT00000064345 |
Mga
|
MGA, MAX dimerization protein |
| chr13_-_94289333 | 3.20 |
ENSRNOT00000078195
|
Pld5
|
phospholipase D family, member 5 |
| chr14_-_28536260 | 3.16 |
ENSRNOT00000059942
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr2_+_223029559 | 3.01 |
ENSRNOT00000045584
|
AABR07013111.1
|
|
| chr12_-_43576754 | 3.00 |
ENSRNOT00000042331
ENSRNOT00000081780 |
Med13l
|
mediator complex subunit 13-like |
| chr3_+_129599353 | 2.98 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
| chr4_+_168689163 | 2.97 |
ENSRNOT00000049848
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr3_-_8955538 | 2.85 |
ENSRNOT00000040570
|
LOC686066
|
similar to 60S ribosomal protein L38 |
| chr4_+_66670618 | 2.82 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr18_+_17403407 | 2.56 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr4_-_114823500 | 2.48 |
ENSRNOT00000089870
|
NEWGENE_1562258
|
INO80 complex subunit B |
| chr16_+_90325304 | 2.37 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr17_-_63879166 | 2.32 |
ENSRNOT00000049391
ENSRNOT00000045183 |
Zmynd11
|
zinc finger, MYND-type containing 11 |
| chr14_-_16276068 | 2.30 |
ENSRNOT00000002876
|
Ccng2
|
cyclin G2 |
| chr18_+_62852303 | 2.30 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr20_+_6356423 | 2.21 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr7_-_60742660 | 2.18 |
ENSRNOT00000086116
|
Mdm2
|
MDM2 proto-oncogene |
| chr13_+_92167643 | 2.17 |
ENSRNOT00000030794
|
Olr1589
|
olfactory receptor 1589 |
| chr18_+_48853418 | 2.15 |
ENSRNOT00000022792
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr14_+_89314176 | 2.14 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr13_-_26769374 | 2.12 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr8_-_55276070 | 2.12 |
ENSRNOT00000041555
|
LOC108351703
|
60S ribosomal protein L27a-like |
| chr2_+_45104305 | 2.00 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr7_+_116357175 | 2.00 |
ENSRNOT00000076790
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr18_+_51523758 | 1.96 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr7_+_144595960 | 1.94 |
ENSRNOT00000021909
|
Hoxc9
|
homeobox C9 |
| chr12_+_13715843 | 1.90 |
ENSRNOT00000042459
|
Actb
|
actin, beta |
| chrX_+_74824504 | 1.86 |
ENSRNOT00000048963
|
LOC680353
|
similar to 60S ribosomal protein L38 |
| chr7_+_70445366 | 1.83 |
ENSRNOT00000045687
|
RGD1565117
|
similar to 40S ribosomal protein S26 |
| chr16_-_32540217 | 1.82 |
ENSRNOT00000015130
|
Hpf1
|
histone PARylation factor 1 |
| chr18_+_47739140 | 1.70 |
ENSRNOT00000082760
|
Sncaip
|
synuclein, alpha interacting protein |
| chr8_+_87211819 | 1.63 |
ENSRNOT00000086093
|
LOC100363289
|
LRRGT00022-like |
| chr20_+_4143321 | 1.58 |
ENSRNOT00000060366
|
Btnl2
|
butyrophilin-like 2 |
| chr8_+_98755104 | 1.56 |
ENSRNOT00000056562
|
Zic4
|
Zic family member 4 |
| chr8_-_108261021 | 1.56 |
ENSRNOT00000040710
|
LOC100912210
|
40S ribosomal protein S25-like |
| chr2_-_111793326 | 1.50 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr10_+_68588789 | 1.45 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr10_-_97756521 | 1.34 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr20_+_27212724 | 1.28 |
ENSRNOT00000032600
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr4_-_51844331 | 1.28 |
ENSRNOT00000003593
|
Gpr37
|
G protein-coupled receptor 37 |
| chr1_-_219395623 | 1.25 |
ENSRNOT00000040261
|
Rpl35a
|
ribosomal protein L35a |
| chr20_+_48212378 | 1.24 |
ENSRNOT00000040519
|
AABR07045454.1
|
|
| chr17_+_18358712 | 1.19 |
ENSRNOT00000001979
|
Nup153
|
nucleoporin 153 |
| chr2_+_76923591 | 1.17 |
ENSRNOT00000042759
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chr17_-_9519595 | 1.09 |
ENSRNOT00000029885
|
Ddx46
|
DEAD-box helicase 46 |
| chr10_-_104522241 | 1.05 |
ENSRNOT00000046010
|
Rps18l1
|
ribosomal protein S18-like 1 |
| chr1_+_255479261 | 1.00 |
ENSRNOT00000079808
|
Tnks2
|
tankyrase 2 |
| chr3_-_55451798 | 0.97 |
ENSRNOT00000008837
|
Spc25
|
SPC25, NDC80 kinetochore complex component |
| chr15_-_19282873 | 0.89 |
ENSRNOT00000075184
|
NEWGENE_1359268
|
Smad nuclear interacting protein 1 |
| chr3_-_20914603 | 0.89 |
ENSRNOT00000049651
|
Olr416
|
olfactory receptor 416 |
| chr3_-_154042099 | 0.87 |
ENSRNOT00000033525
|
Blcap
|
bladder cancer associated protein |
| chr1_+_190852985 | 0.86 |
ENSRNOT00000040963
|
Polr3e
|
RNA polymerase III subunit E |
| chr4_-_152883210 | 0.85 |
ENSRNOT00000056200
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr1_+_72756809 | 0.80 |
ENSRNOT00000082423
|
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1 |
| chr2_-_77784522 | 0.80 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr6_+_134844676 | 0.76 |
ENSRNOT00000007595
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_76256129 | 0.70 |
ENSRNOT00000072741
|
LOC100912605
|
olfactory receptor 5D14-like |
| chr15_-_30147793 | 0.70 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr1_-_211923929 | 0.68 |
ENSRNOT00000054887
|
Nkx6-2
|
NK6 homeobox 2 |
| chr10_-_35175647 | 0.56 |
ENSRNOT00000078960
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr5_-_100977902 | 0.56 |
ENSRNOT00000077429
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr18_+_35850132 | 0.55 |
ENSRNOT00000067337
ENSRNOT00000075841 |
LOC103690064
|
m7GpppN-mRNA hydrolase-like |
| chr1_-_91588609 | 0.49 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr20_+_1764794 | 0.41 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr18_-_27374603 | 0.39 |
ENSRNOT00000027244
|
Nme5
|
NME/NM23 family member 5 |
| chr4_+_89536647 | 0.33 |
ENSRNOT00000058581
|
Tigd2
|
tigger transposable element derived 2 |
| chr13_+_92224971 | 0.25 |
ENSRNOT00000004674
|
Olr1593
|
olfactory receptor 1593 |
| chr20_+_1749716 | 0.25 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr2_-_149754776 | 0.23 |
ENSRNOT00000035057
|
RGD1559622
|
similar to hypothetical protein C130079G13 |
| chr11_-_782954 | 0.18 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr1_+_65781547 | 0.11 |
ENSRNOT00000026204
|
Zfp329
|
zinc finger protein 329 |
| chr15_+_47373120 | 0.10 |
ENSRNOT00000070815
|
Rp1l1
|
RP1 like 1 |
| chr13_+_96122120 | 0.07 |
ENSRNOT00000087964
|
Desi2
|
desumoylating isopeptidase 2 |
| chr5_+_116420690 | 0.07 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr19_-_53754602 | 0.07 |
ENSRNOT00000035651
|
Fam92b
|
family with sequence similarity 92, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 1.8 | 5.4 | GO:1901580 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.7 | 8.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 4.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.3 | 9.4 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 1.1 | 3.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.0 | 3.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.9 | 6.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.9 | 6.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.8 | 1.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.7 | 2.2 | GO:1904404 | traversing start control point of mitotic cell cycle(GO:0007089) response to formaldehyde(GO:1904404) |
| 0.7 | 2.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.7 | 3.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.6 | 10.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.6 | 1.8 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.6 | 4.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.6 | 8.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.5 | 7.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 2.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.5 | 3.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 3.5 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.4 | 2.1 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.3 | 0.7 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 3.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 2.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.3 | 2.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.3 | 3.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.3 | 4.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.3 | 2.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 4.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 6.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 3.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.0 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.1 | 0.6 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 3.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 2.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.9 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 1.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.3 | GO:0046329 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 9.1 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 4.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 3.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 8.6 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.9 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.9 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 7.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 11.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 10.9 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.8 | 4.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 2.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 3.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.4 | 3.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 11.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 3.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.4 | 7.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 6.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 12.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 3.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 12.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 1.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 1.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 3.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 9.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 10.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 12.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.3 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 8.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 17.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.3 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 1.1 | 5.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.1 | 6.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.7 | 2.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 3.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 2.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.6 | 4.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.5 | 10.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 9.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 2.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 10.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.4 | 2.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 2.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 4.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 7.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 3.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 3.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 13.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 8.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 7.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 1.9 | GO:0098918 | structural constituent of synapse(GO:0098918) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 8.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 2.8 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 3.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 10.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 6.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 8.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 12.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 2.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 7.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 6.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 2.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 8.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 5.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 8.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 6.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.5 | 6.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 7.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.4 | 10.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 2.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 3.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 9.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 4.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 3.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |