Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
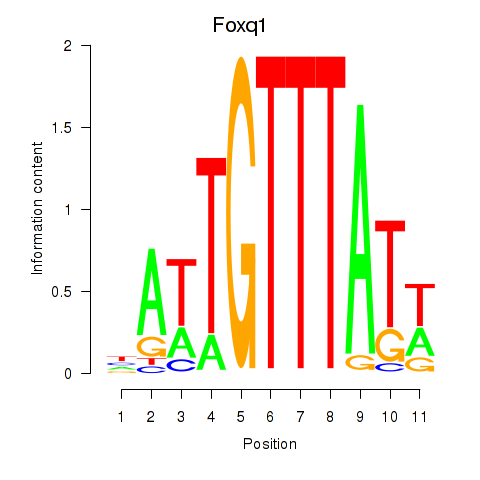
Results for Foxq1
Z-value: 0.77
Transcription factors associated with Foxq1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxq1
|
ENSRNOG00000021752 | Foxq1 |
Activity profile of Foxq1 motif
Sorted Z-values of Foxq1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32973695 | 24.49 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr3_-_72219246 | 22.37 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr2_-_199971965 | 19.98 |
ENSRNOT00000087026
|
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr10_-_34439470 | 19.71 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr8_-_73194837 | 19.41 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr14_+_99529284 | 17.51 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr16_-_10802512 | 16.17 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr20_-_18060661 | 16.15 |
ENSRNOT00000070925
|
AABR07044711.1
|
|
| chr2_-_172361779 | 15.10 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr18_+_30592794 | 15.09 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr3_-_123607352 | 14.38 |
ENSRNOT00000051404
ENSRNOT00000028854 |
Adam33
|
ADAM metallopeptidase domain 33 |
| chrX_+_53053609 | 14.36 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chrX_-_142248369 | 14.19 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr18_+_30527705 | 13.95 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr4_+_139670092 | 13.77 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr16_+_81593532 | 13.45 |
ENSRNOT00000026370
|
NEWGENE_1582994
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr1_+_164380577 | 12.93 |
ENSRNOT00000055321
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr17_+_47721977 | 12.36 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr5_-_102786331 | 11.97 |
ENSRNOT00000086635
|
Bnc2
|
basonuclin 2 |
| chr4_-_115239723 | 11.87 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_-_165967069 | 11.36 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr20_+_29655226 | 11.08 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chrX_-_74968405 | 10.62 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chrX_+_15669191 | 10.33 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chr2_-_233743866 | 9.81 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr17_-_13393243 | 9.69 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr11_-_28527890 | 9.48 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr6_-_95998529 | 9.26 |
ENSRNOT00000050138
|
Six4
|
SIX homeobox 4 |
| chrX_+_121612952 | 8.98 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr18_+_30562178 | 8.33 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chrM_+_5323 | 8.23 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr4_-_159399634 | 8.13 |
ENSRNOT00000089193
|
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr2_-_53300404 | 7.49 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr6_+_76677213 | 7.45 |
ENSRNOT00000076760
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr18_+_65155685 | 6.86 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr17_-_15484269 | 6.85 |
ENSRNOT00000020648
|
Omd
|
osteomodulin |
| chr16_-_74710704 | 6.85 |
ENSRNOT00000016859
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chr5_-_169160987 | 6.58 |
ENSRNOT00000055487
|
Thap3
|
THAP domain containing 3 |
| chrX_-_77559348 | 6.51 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr3_+_134440195 | 6.47 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr1_-_216744066 | 6.26 |
ENSRNOT00000087961
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_99894762 | 6.24 |
ENSRNOT00000081768
|
Plscr4
|
phospholipid scramblase 4 |
| chr1_+_42121636 | 6.00 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr4_-_55407922 | 5.71 |
ENSRNOT00000031714
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr4_-_159287193 | 5.71 |
ENSRNOT00000080139
|
Kcna6
|
potassium voltage-gated channel subfamily A member 6 |
| chr14_-_6900733 | 5.55 |
ENSRNOT00000061224
|
Dmp1
|
dentin matrix acidic phosphoprotein 1 |
| chr7_+_142776252 | 5.20 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr7_-_138707221 | 5.05 |
ENSRNOT00000009199
|
Amigo2
|
adhesion molecule with Ig like domain 2 |
| chr17_+_49417067 | 4.77 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr18_+_57654290 | 4.58 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chrX_+_83678339 | 4.50 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr8_+_49621568 | 4.46 |
ENSRNOT00000021949
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr12_+_29921443 | 4.37 |
ENSRNOT00000001190
|
Sbds
|
SBDS ribosome assembly guanine nucleotide exchange factor |
| chr1_-_167911961 | 4.29 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr7_+_142776580 | 4.25 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr11_-_28979169 | 4.22 |
ENSRNOT00000061601
|
AABR07033580.1
|
|
| chr7_-_144272578 | 4.02 |
ENSRNOT00000020676
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr20_+_5094468 | 3.93 |
ENSRNOT00000078576
|
Abhd16a
|
abhydrolase domain containing 16A |
| chr2_+_188087486 | 3.91 |
ENSRNOT00000027455
|
Rit1
|
Ras-like without CAAX 1 |
| chr1_+_259958310 | 3.89 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr7_-_9465527 | 3.80 |
ENSRNOT00000044882
|
Olr1069
|
olfactory receptor 1069 |
| chr16_-_8350067 | 3.79 |
ENSRNOT00000026932
|
LOC100362432
|
mitochondrial import inner membrane translocase subunit Tim23-like |
| chr4_+_121899292 | 3.74 |
ENSRNOT00000077949
|
Vom1r97
|
vomeronasal 1 receptor 97 |
| chr4_+_161720501 | 3.69 |
ENSRNOT00000008196
ENSRNOT00000055886 |
Nrip2
|
nuclear receptor interacting protein 2 |
| chr11_+_86421106 | 3.46 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr3_-_177078594 | 3.35 |
ENSRNOT00000020695
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr8_-_122311431 | 3.33 |
ENSRNOT00000033126
|
Fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr17_-_77527894 | 3.19 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr1_-_101773508 | 3.16 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr1_-_263431290 | 3.09 |
ENSRNOT00000022633
|
Slc25a28
|
solute carrier family 25 member 28 |
| chr3_-_2845593 | 3.04 |
ENSRNOT00000067479
|
Tmem141
|
transmembrane protein 141 |
| chr3_-_78526022 | 3.00 |
ENSRNOT00000082898
|
Olr709
|
olfactory receptor 709 |
| chr7_-_143167772 | 3.00 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr2_+_257295193 | 2.98 |
ENSRNOT00000052249
|
AABR07013798.1
|
|
| chr13_-_49487892 | 2.85 |
ENSRNOT00000044972
|
Nfasc
|
neurofascin |
| chr1_-_253000760 | 2.83 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr3_+_147511406 | 2.69 |
ENSRNOT00000082438
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr5_+_158090173 | 2.69 |
ENSRNOT00000088766
ENSRNOT00000079516 ENSRNOT00000092026 |
Tas1r2
|
taste 1 receptor member 2 |
| chr3_+_76842898 | 2.66 |
ENSRNOT00000085537
|
Olr636
|
olfactory receptor 636 |
| chr8_-_61519507 | 2.54 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr16_+_48513432 | 2.44 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr14_+_34455934 | 2.43 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chr15_-_18632536 | 2.41 |
ENSRNOT00000061123
|
Rpp14
|
ribonuclease P/MRP 14 subunit |
| chr13_+_26970660 | 2.39 |
ENSRNOT00000083145
|
Serpinb12
|
serpin family B member 12 |
| chr10_+_66732390 | 2.37 |
ENSRNOT00000089538
|
Nf1
|
neurofibromin 1 |
| chr4_+_163293724 | 2.36 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr11_-_11585078 | 2.35 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_-_2832697 | 2.19 |
ENSRNOT00000065410
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr2_-_244370983 | 2.11 |
ENSRNOT00000021458
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr7_-_3707226 | 2.09 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr5_+_113725717 | 2.07 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr4_-_9921610 | 2.07 |
ENSRNOT00000017018
|
Pmpcb
|
peptidase, mitochondrial processing beta subunit |
| chr6_-_123405246 | 2.04 |
ENSRNOT00000083665
|
Foxn3
|
forkhead box N3 |
| chrX_-_143274180 | 2.03 |
ENSRNOT00000051550
|
Mcf2
|
MCF.2 cell line derived transforming sequence |
| chr4_-_165747201 | 1.98 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr7_-_66172360 | 1.96 |
ENSRNOT00000005537
|
Fam19a2
|
family with sequence similarity 19 member A2, C-C motif chemokine like |
| chr13_+_74456487 | 1.94 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr7_+_3900228 | 1.88 |
ENSRNOT00000071350
|
Olr921
|
olfactory receptor 921 |
| chr10_-_60573129 | 1.84 |
ENSRNOT00000083428
|
Olr1499
|
olfactory receptor 1499 |
| chr11_-_45616429 | 1.82 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
| chr3_-_160038078 | 1.81 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr14_+_15949171 | 1.79 |
ENSRNOT00000041124
|
LOC100363333
|
PRAME family member 12 |
| chr2_-_210943620 | 1.78 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr3_+_78009498 | 1.72 |
ENSRNOT00000042640
|
Olr686
|
olfactory receptor 686 |
| chr20_-_43932361 | 1.72 |
ENSRNOT00000091030
|
Rfpl4b
|
ret finger protein-like 4B |
| chr5_-_60658521 | 1.70 |
ENSRNOT00000092866
ENSRNOT00000093024 ENSRNOT00000016875 |
Tomm5
Fbxo10
|
translocase of outer mitochondrial membrane 5 F-box protein 10 |
| chr7_-_130176696 | 1.69 |
ENSRNOT00000051347
|
Dennd6b
|
DENN domain containing 6B |
| chr8_+_117282390 | 1.63 |
ENSRNOT00000074772
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr1_+_86103298 | 1.61 |
ENSRNOT00000014883
|
Vom2r10
|
vomeronasal 2 receptor, 10 |
| chr7_+_13378338 | 1.61 |
ENSRNOT00000042747
|
Olr1073
|
olfactory receptor 1073 |
| chr3_-_77961992 | 1.56 |
ENSRNOT00000089105
|
AC132028.2
|
|
| chr13_-_91735361 | 1.47 |
ENSRNOT00000058090
|
Fcer1a
|
Fc fragment of IgE receptor Ia |
| chr8_+_56585396 | 1.36 |
ENSRNOT00000085316
ENSRNOT00000034924 |
Rdx
|
radixin |
| chr3_+_147511563 | 1.28 |
ENSRNOT00000006828
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr10_-_44972367 | 1.23 |
ENSRNOT00000048778
|
LOC100910876
|
olfactory receptor 2AK2-like |
| chr7_-_13461405 | 1.20 |
ENSRNOT00000083574
|
Olr1077
|
olfactory receptor 1077 |
| chr5_-_16747222 | 1.19 |
ENSRNOT00000011602
|
Mos
|
Moloney sarcoma oncogene |
| chr1_+_64126882 | 1.16 |
ENSRNOT00000091742
|
Leng1
|
leukocyte receptor cluster member 1 |
| chr1_+_244630377 | 1.15 |
ENSRNOT00000016740
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_6803318 | 1.12 |
ENSRNOT00000084203
|
Olr954
|
olfactory receptor 954 |
| chr8_+_131998778 | 1.12 |
ENSRNOT00000040625
|
Zfp105
|
zinc finger protein 105 |
| chr13_-_78885464 | 1.08 |
ENSRNOT00000003828
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr8_+_43763859 | 1.04 |
ENSRNOT00000080504
|
Olr1328
|
olfactory receptor 1328 |
| chr3_+_77337698 | 1.02 |
ENSRNOT00000041690
|
LOC103690075
|
olfactory receptor 4P4-like |
| chr3_-_78409263 | 1.00 |
ENSRNOT00000041001
|
Olr704
|
olfactory receptor 704 |
| chr19_-_21970103 | 0.92 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr2_-_256154584 | 0.90 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr3_+_38559914 | 0.90 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr11_+_62584959 | 0.87 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr7_-_139835876 | 0.84 |
ENSRNOT00000014258
|
Olr1877
|
olfactory receptor 1877 |
| chr4_+_71512695 | 0.81 |
ENSRNOT00000037220
|
Tas2r139
|
taste receptor, type 2, member 139 |
| chr2_-_183558408 | 0.76 |
ENSRNOT00000082077
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr12_-_11176106 | 0.68 |
ENSRNOT00000090073
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr3_+_77072410 | 0.66 |
ENSRNOT00000078540
|
Olr649
|
olfactory receptor 649 |
| chr3_-_78555530 | 0.65 |
ENSRNOT00000089672
|
Olr711
|
olfactory receptor 711 |
| chr3_-_98092131 | 0.64 |
ENSRNOT00000006512
|
Fshb
|
follicle stimulating hormone beta subunit |
| chr4_+_168615890 | 0.61 |
ENSRNOT00000009324
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr3_+_75771589 | 0.61 |
ENSRNOT00000007665
|
Olr598
|
olfactory receptor 598 |
| chr10_-_91581232 | 0.54 |
ENSRNOT00000066453
ENSRNOT00000068686 |
AABR07030520.1
|
|
| chr1_+_89785552 | 0.46 |
ENSRNOT00000066292
ENSRNOT00000052376 |
Scgb1b24
|
secretoglobin, family 1B, member 24 |
| chr15_+_4882947 | 0.44 |
ENSRNOT00000049255
ENSRNOT00000092002 |
RGD1304704
|
similar to Hypothetical protein CGI-99 |
| chr12_-_19254527 | 0.42 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr15_+_33108671 | 0.39 |
ENSRNOT00000015468
|
Lrp10
|
LDL receptor related protein 10 |
| chr12_-_11175917 | 0.37 |
ENSRNOT00000079573
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr2_+_211320420 | 0.27 |
ENSRNOT00000027504
|
RGD1309139
|
similar to CG5435-PA |
| chr3_-_76368909 | 0.22 |
ENSRNOT00000043599
|
Olr614
|
olfactory receptor 614 |
| chr5_-_16995304 | 0.18 |
ENSRNOT00000061780
|
Sdr16c6
|
short chain dehydrogenase/reductase family 16C, member 6 |
| chr9_+_65177511 | 0.16 |
ENSRNOT00000048027
|
Aox2
|
aldehyde oxidase 2 |
| chr3_-_13896838 | 0.10 |
ENSRNOT00000025399
|
Fbxw2
|
F-box and WD repeat domain containing 2 |
| chr4_+_147929863 | 0.07 |
ENSRNOT00000015427
|
LOC680385
|
similar to Sjogren syndrome antigen B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxq1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0048372 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 4.8 | 14.4 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 3.5 | 17.5 | GO:0070343 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 2.4 | 11.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.3 | 9.3 | GO:0071699 | olfactory placode formation(GO:0030910) myotome development(GO:0061055) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 2.1 | 10.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 2.0 | 14.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 2.0 | 9.8 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.8 | 19.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.4 | 4.3 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 1.4 | 11.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.3 | 4.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 1.1 | 5.6 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 1.1 | 9.5 | GO:0035912 | dorsal aorta morphogenesis(GO:0035912) |
| 1.0 | 3.9 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.9 | 2.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.8 | 3.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.8 | 7.5 | GO:0046449 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) |
| 0.8 | 12.9 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.7 | 2.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 15.1 | GO:0001553 | luteinization(GO:0001553) estrogen metabolic process(GO:0008210) |
| 0.7 | 8.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.6 | 4.4 | GO:0042256 | mature ribosome assembly(GO:0042256) bone marrow development(GO:0048539) |
| 0.6 | 2.4 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.6 | 6.9 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.6 | 5.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.6 | 9.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 2.7 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 0.5 | 8.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 1.5 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.5 | 2.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 | 1.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 3.2 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.4 | 12.0 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.4 | 1.2 | GO:1902102 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.4 | 1.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 2.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 16.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.3 | 42.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 1.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 4.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 2.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.2 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 2.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 4.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.2 | 4.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 3.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 2.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 4.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 11.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 3.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 2.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 2.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 6.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 1.6 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 2.8 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 7.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 6.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 3.8 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.0 | 0.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 20.0 | GO:0043623 | cellular protein complex assembly(GO:0043623) |
| 0.0 | 2.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 11.4 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 3.7 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 3.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.6 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 18.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.0 | GO:0016358 | dendrite development(GO:0016358) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.5 | 7.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.0 | 19.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.8 | 22.4 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 11.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.8 | 14.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.7 | 9.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 12.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 4.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 2.8 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 2.7 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.5 | 8.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 9.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 2.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 3.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.4 | 15.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.4 | 7.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 4.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 14.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 2.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 8.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 16.2 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 37.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 15.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.1 | 5.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 5.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 29.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 6.3 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 25.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 2.3 | 6.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.9 | 7.5 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.6 | 9.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.4 | 5.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.3 | 4.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 1.2 | 22.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.1 | 3.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.7 | 14.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.7 | 2.7 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.7 | 3.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 6.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 9.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 3.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.4 | 1.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 3.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.3 | 2.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 14.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 3.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 2.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 8.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.3 | 8.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 4.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 2.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 16.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 5.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 7.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 2.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 19.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 3.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.8 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 3.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 16.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 4.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 9.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 3.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 23.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 23.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.5 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 3.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 6.8 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 17.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 9.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 9.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 16.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 11.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 13.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 13.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 6.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 16.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 2.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 5.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.6 | 16.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 6.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 26.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.4 | 8.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 7.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 4.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 9.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 13.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 6.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 8.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |