Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
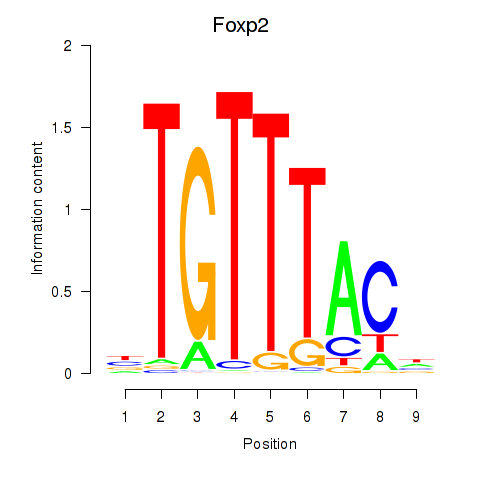
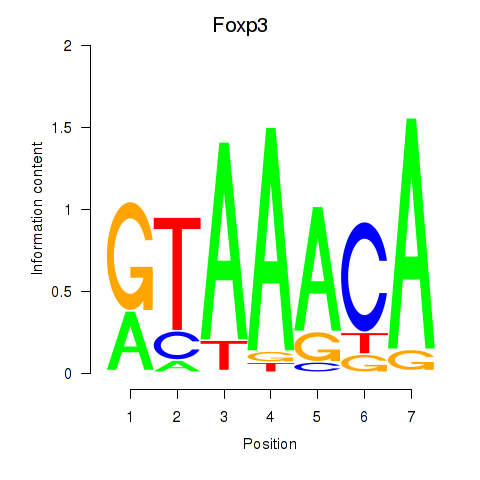
Results for Foxp2_Foxp3
Z-value: 0.86
Transcription factors associated with Foxp2_Foxp3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxp2
|
ENSRNOG00000054508 | forkhead box P2 |
|
Foxp3
|
ENSRNOG00000011702 | forkhead box P3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxp3 | rn6_v1_chrX_-_15761932_15761932 | 0.28 | 4.7e-07 | Click! |
| Foxp2 | rn6_v1_chr4_+_41364441_41364441 | 0.23 | 4.4e-05 | Click! |
Activity profile of Foxp2_Foxp3 motif
Sorted Z-values of Foxp2_Foxp3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_103395511 | 26.13 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr10_+_96639924 | 21.80 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr4_-_30556814 | 20.82 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr4_-_82289672 | 19.96 |
ENSRNOT00000036394
|
Hoxa10
|
homeobox A10 |
| chr7_+_58814805 | 19.87 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr7_+_74047814 | 19.12 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr6_+_129399468 | 17.77 |
ENSRNOT00000071735
ENSRNOT00000074621 |
Bdkrb2
|
bradykinin receptor B2 |
| chr4_-_82295470 | 17.48 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr5_+_124300477 | 16.85 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr4_-_82209933 | 16.85 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr14_-_19072677 | 16.79 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr5_-_88629491 | 14.69 |
ENSRNOT00000058906
|
Tle1
|
transducin like enhancer of split 1 |
| chr4_+_169147243 | 14.68 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr2_-_233743866 | 13.86 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr5_+_152533349 | 13.54 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr4_-_115239723 | 13.37 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_-_108167185 | 13.16 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr4_+_169161585 | 12.92 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr20_-_54517709 | 12.68 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr15_-_28313841 | 12.10 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr13_-_98480419 | 12.10 |
ENSRNOT00000086306
|
Coq8a
|
coenzyme Q8A |
| chr8_-_49109981 | 11.89 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr5_-_101166651 | 11.83 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr10_+_56662242 | 11.63 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr13_-_1946508 | 11.47 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr1_+_225184939 | 11.24 |
ENSRNOT00000079456
|
Ahnak
|
AHNAK nucleoprotein |
| chr7_-_29171783 | 10.90 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr7_+_144577465 | 10.87 |
ENSRNOT00000021647
|
Hoxc10
|
homeo box C10 |
| chr2_-_204254699 | 10.74 |
ENSRNOT00000021487
|
Mab21l3
|
mab-21 like 3 |
| chr4_+_83713666 | 10.54 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr7_-_97605113 | 10.31 |
ENSRNOT00000089734
|
AABR07058000.1
|
|
| chr7_-_138483612 | 10.27 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_+_25349928 | 10.21 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chrX_-_111887906 | 10.06 |
ENSRNOT00000085118
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr1_-_267203986 | 9.95 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr2_-_98610368 | 9.69 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr12_+_9446940 | 9.44 |
ENSRNOT00000074791
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr10_-_74298599 | 9.39 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr1_+_250426158 | 9.29 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr18_+_3887419 | 9.28 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr2_-_53300404 | 9.24 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr4_+_154309426 | 9.18 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr18_+_29352749 | 8.86 |
ENSRNOT00000025137
|
Slc4a9
|
solute carrier family 4 member 9 |
| chr7_-_119689938 | 8.81 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr17_+_12310214 | 8.79 |
ENSRNOT00000015786
|
Auh
|
AU RNA binding methylglutaconyl-CoA hydratase |
| chr7_+_99954492 | 8.72 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr17_+_32973695 | 8.63 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr10_+_11393103 | 8.44 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr8_-_104593625 | 8.41 |
ENSRNOT00000016625
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr6_-_127534247 | 8.38 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr4_+_62380914 | 8.37 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr10_+_56824505 | 8.20 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr9_+_43093138 | 7.99 |
ENSRNOT00000021592
|
Cnnm3
|
cyclin and CBS domain divalent metal cation transport mediator 3 |
| chr1_-_48360131 | 7.96 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr8_+_65733400 | 7.93 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chrX_+_15669191 | 7.92 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chr15_+_34216833 | 7.83 |
ENSRNOT00000025260
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr4_+_44774741 | 7.74 |
ENSRNOT00000086902
|
Met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr12_-_1816414 | 7.70 |
ENSRNOT00000041155
ENSRNOT00000067448 |
Insr
|
insulin receptor |
| chr1_-_131460473 | 7.60 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr18_+_61258911 | 7.38 |
ENSRNOT00000082403
|
Zfp532
|
zinc finger protein 532 |
| chr1_-_31055453 | 7.36 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr3_+_126335863 | 7.36 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr1_+_220114228 | 7.33 |
ENSRNOT00000026718
|
Ctsf
|
cathepsin F |
| chr15_-_37983882 | 7.28 |
ENSRNOT00000087978
|
Lats2
|
large tumor suppressor kinase 2 |
| chrX_+_119390013 | 7.20 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr14_-_60964324 | 7.17 |
ENSRNOT00000005155
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr11_-_61475681 | 7.15 |
ENSRNOT00000093249
|
Usf3
|
upstream transcription factor family member 3 |
| chr9_-_52238564 | 7.12 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr3_+_11114551 | 7.07 |
ENSRNOT00000013507
|
Plpp7
|
phospholipid phosphatase 7 |
| chr1_+_100845563 | 7.03 |
ENSRNOT00000027478
|
Akt1s1
|
AKT1 substrate 1 |
| chr3_+_101791337 | 6.99 |
ENSRNOT00000046937
|
Slc5a12
|
solute carrier family 5 member 12 |
| chr3_-_57957346 | 6.88 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr18_+_65155685 | 6.69 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr1_+_243375981 | 6.65 |
ENSRNOT00000080689
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr11_-_45124423 | 6.55 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr15_+_34234755 | 6.54 |
ENSRNOT00000059987
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr8_+_50559126 | 6.52 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr7_+_3900228 | 6.46 |
ENSRNOT00000071350
|
Olr921
|
olfactory receptor 921 |
| chr4_+_115416580 | 6.43 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr2_+_185393441 | 6.42 |
ENSRNOT00000015802
|
Sh3d19
|
SH3 domain containing 19 |
| chr8_-_50245838 | 6.34 |
ENSRNOT00000035310
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr13_+_50103189 | 6.31 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr8_+_57886168 | 6.31 |
ENSRNOT00000039336
|
Exph5
|
exophilin 5 |
| chr7_+_143749221 | 6.27 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr4_+_83391283 | 6.27 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr5_-_124403195 | 6.12 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr3_-_72219246 | 6.01 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr19_-_52206310 | 6.01 |
ENSRNOT00000087886
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr1_+_260798239 | 6.01 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr15_+_24942218 | 5.93 |
ENSRNOT00000016629
|
Peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr13_+_80125391 | 5.92 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr1_+_107344904 | 5.84 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr4_+_41364441 | 5.84 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr10_-_87500919 | 5.83 |
ENSRNOT00000084798
|
Krtap1-5
|
keratin associated protein 1-5 |
| chr14_-_66978499 | 5.78 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr9_+_74124016 | 5.76 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr17_-_22143324 | 5.73 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr5_+_162808646 | 5.69 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr10_+_98706960 | 5.69 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr3_+_72460889 | 5.67 |
ENSRNOT00000012209
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr6_+_1147012 | 5.58 |
ENSRNOT00000006360
|
Vit
|
vitrin |
| chr10_+_56662561 | 5.58 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr4_+_62221011 | 5.54 |
ENSRNOT00000041264
|
Cald1
|
caldesmon 1 |
| chr8_-_50246656 | 5.51 |
ENSRNOT00000024240
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr5_-_28131133 | 5.37 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr5_-_60559533 | 5.36 |
ENSRNOT00000092899
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr4_+_139670092 | 5.33 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chrX_-_15627235 | 5.31 |
ENSRNOT00000013369
|
Wdr45
|
WD repeat domain 45 |
| chr9_+_64745051 | 5.30 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_198016898 | 5.27 |
ENSRNOT00000025523
|
Car14
|
carbonic anhydrase 14 |
| chr2_+_222021103 | 5.21 |
ENSRNOT00000086125
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr2_+_155718359 | 5.19 |
ENSRNOT00000077973
|
Kcnab1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr5_-_33892462 | 5.19 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr2_+_198683159 | 5.15 |
ENSRNOT00000028793
|
Txnip
|
thioredoxin interacting protein |
| chr1_-_236900904 | 5.15 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr4_+_62220736 | 5.13 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr1_+_141218095 | 5.04 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr10_-_40296470 | 5.03 |
ENSRNOT00000086456
|
Tnip1
|
TNFAIP3 interacting protein 1 |
| chrX_-_25590048 | 5.01 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr1_-_206282575 | 5.00 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr1_+_221236773 | 4.96 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr15_+_34552410 | 4.96 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr20_+_18780605 | 4.89 |
ENSRNOT00000000754
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr2_-_187909394 | 4.81 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chrX_+_65566047 | 4.74 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr4_-_170860225 | 4.71 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr6_+_12362813 | 4.71 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr6_+_43234526 | 4.68 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr18_+_16146447 | 4.66 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr1_-_263762785 | 4.61 |
ENSRNOT00000018221
|
Cpn1
|
carboxypeptidase N subunit 1 |
| chrX_+_157759624 | 4.52 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr8_-_125645898 | 4.49 |
ENSRNOT00000036672
|
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr7_+_141053876 | 4.48 |
ENSRNOT00000084674
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chrM_-_14061 | 4.40 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr18_+_867048 | 4.39 |
ENSRNOT00000065494
|
Colec12
|
collectin sub-family member 12 |
| chr1_+_240908483 | 4.37 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr15_-_33051129 | 4.36 |
ENSRNOT00000014391
|
Slc7a7
|
solute carrier family 7 member 7 |
| chr10_-_78155999 | 4.35 |
ENSRNOT00000092852
|
Stxbp4
|
syntaxin binding protein 4 |
| chr3_+_117421604 | 4.35 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr13_-_80775230 | 4.33 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr1_+_243662823 | 4.33 |
ENSRNOT00000067372
|
Dmrt2
|
doublesex and mab-3 related transcription factor 2 |
| chr1_-_252550394 | 4.31 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_-_144269486 | 4.29 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chrX_+_78042859 | 4.28 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr6_-_122721496 | 4.28 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr13_+_89597138 | 4.23 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr5_+_48374096 | 4.23 |
ENSRNOT00000010172
|
Gabrr1
|
gamma-aminobutyric acid type A receptor rho 1 subunit |
| chr3_-_161299024 | 4.15 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr2_-_192671059 | 4.13 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr17_-_15519060 | 4.10 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr1_-_100845392 | 4.06 |
ENSRNOT00000027436
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr3_-_119330345 | 4.04 |
ENSRNOT00000077398
ENSRNOT00000079191 |
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_+_209161047 | 4.00 |
ENSRNOT00000023903
|
Dram2
|
DNA-damage regulated autophagy modulator 2 |
| chrX_+_111122552 | 4.00 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr15_+_59678165 | 3.99 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr9_-_38196273 | 3.98 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr15_+_55126953 | 3.96 |
ENSRNOT00000021430
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr8_+_85417838 | 3.92 |
ENSRNOT00000012257
|
Ick
|
intestinal cell kinase |
| chr1_+_68436593 | 3.89 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chrX_-_118513061 | 3.89 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr4_+_154423209 | 3.88 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr11_-_32550539 | 3.88 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chrX_+_143097525 | 3.87 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr1_+_137014272 | 3.85 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr3_-_2453933 | 3.80 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr1_+_100473643 | 3.77 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr1_-_277181345 | 3.74 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr6_-_51257625 | 3.73 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr1_+_83003841 | 3.72 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr1_-_275876329 | 3.72 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr3_+_60024013 | 3.71 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr18_+_27558089 | 3.69 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr1_+_247228061 | 3.64 |
ENSRNOT00000020809
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr20_+_26893016 | 3.64 |
ENSRNOT00000082430
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr16_+_29674793 | 3.62 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr15_+_41937880 | 3.62 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr15_-_9086282 | 3.61 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr3_+_5519990 | 3.60 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr9_+_61720583 | 3.58 |
ENSRNOT00000020536
|
Mob4
|
MOB family member 4, phocein |
| chr1_-_127292090 | 3.55 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr7_+_59326518 | 3.53 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr15_+_34234193 | 3.48 |
ENSRNOT00000077584
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_16036185 | 3.47 |
ENSRNOT00000047899
|
LOC108348036
|
olfactory receptor 6C2-like |
| chr4_-_15505362 | 3.47 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr14_+_18983853 | 3.43 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr11_+_74057361 | 3.40 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr1_-_167700332 | 3.39 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr17_-_21739408 | 3.37 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr1_+_82419947 | 3.36 |
ENSRNOT00000027964
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr13_-_97838228 | 3.36 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr17_-_1093873 | 3.28 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr14_-_72025137 | 3.27 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr6_-_3355339 | 3.24 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr7_+_93975451 | 3.24 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr10_-_102423084 | 3.20 |
ENSRNOT00000065145
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr2_+_85305225 | 3.15 |
ENSRNOT00000015904
|
Tas2r119
|
taste receptor, type 2, member 119 |
| chr9_-_54327958 | 3.15 |
ENSRNOT00000019465
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr10_+_105393072 | 3.14 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr15_-_50425900 | 3.13 |
ENSRNOT00000058715
|
Adam28
|
ADAM metallopeptidase domain 28 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxp2_Foxp3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.1 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 5.5 | 21.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 5.4 | 16.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 5.2 | 20.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 4.4 | 17.8 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 4.0 | 12.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 3.6 | 10.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 3.5 | 17.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 2.8 | 13.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 2.7 | 13.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.7 | 10.8 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 2.6 | 7.8 | GO:0006116 | NADH oxidation(GO:0006116) |
| 2.5 | 7.6 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 2.5 | 7.4 | GO:0060128 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 2.4 | 7.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 2.4 | 7.2 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 2.3 | 9.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 2.3 | 9.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 2.3 | 11.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 2.2 | 8.7 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.1 | 2.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 2.0 | 11.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.9 | 7.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.9 | 5.8 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.9 | 5.7 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 1.8 | 7.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.7 | 5.2 | GO:0046127 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 1.7 | 6.9 | GO:0089712 | malate-aspartate shuttle(GO:0043490) L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 1.7 | 6.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.6 | 6.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.5 | 27.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.5 | 16.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.5 | 4.5 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.5 | 4.4 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 1.4 | 4.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 1.4 | 5.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.4 | 2.8 | GO:0072185 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 1.4 | 13.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.3 | 8.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.3 | 9.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 1.3 | 7.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.2 | 48.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 1.2 | 10.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.2 | 5.8 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 1.1 | 4.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 1.1 | 3.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 1.1 | 4.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 1.1 | 4.3 | GO:0061055 | myotome development(GO:0061055) |
| 1.1 | 5.3 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 1.0 | 3.1 | GO:1904531 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) glomerular visceral epithelial cell apoptotic process(GO:1903210) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) |
| 1.0 | 8.0 | GO:0015871 | choline transport(GO:0015871) |
| 1.0 | 3.0 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 1.0 | 5.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.0 | 16.8 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 1.0 | 3.9 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 1.0 | 3.8 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 1.0 | 2.9 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.9 | 4.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.9 | 2.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.9 | 4.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.9 | 2.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.9 | 2.6 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.9 | 3.5 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.9 | 4.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.8 | 2.5 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.8 | 8.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.8 | 3.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.8 | 3.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.8 | 7.0 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.8 | 12.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.8 | 3.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.7 | 5.8 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.7 | 5.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 12.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.7 | 13.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.7 | 2.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.7 | 8.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.7 | 3.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.7 | 2.0 | GO:1905244 | negative regulation of long term synaptic depression(GO:1900453) regulation of intracellular transport of viral material(GO:1901252) regulation of modification of synaptic structure(GO:1905244) |
| 0.7 | 4.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.7 | 3.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.7 | 5.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.6 | 2.6 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.6 | 2.6 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.6 | 1.9 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.6 | 3.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.6 | 1.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.6 | 6.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.6 | 2.5 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.6 | 3.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.6 | 3.6 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.6 | 1.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.6 | 11.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.6 | 2.3 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.6 | 1.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.6 | 6.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.6 | 2.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.6 | 3.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 7.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.5 | 5.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.5 | 3.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.5 | 2.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 1.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.5 | 1.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.5 | 4.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.5 | 6.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 2.3 | GO:0060762 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 6.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.5 | 5.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 8.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.4 | 1.8 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 3.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 3.9 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.4 | 1.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 1.2 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.4 | 3.1 | GO:0015679 | plasma membrane copper ion transport(GO:0015679) |
| 0.4 | 19.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 5.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.4 | 1.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 | 2.3 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.4 | 1.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.4 | 7.8 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.4 | 1.5 | GO:0070627 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.4 | 2.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 2.1 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.4 | 3.5 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.4 | 3.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 2.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 1.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.3 | 5.4 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.3 | 1.3 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.3 | 4.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 2.6 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 2.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 1.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.3 | 4.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 0.9 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.3 | 5.3 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.3 | 1.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 8.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 3.6 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.3 | 0.9 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.3 | 6.2 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.3 | 0.9 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.3 | 0.9 | GO:2000853 | negative regulation of corticosterone secretion(GO:2000853) |
| 0.3 | 6.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.3 | 4.8 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.3 | 1.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.3 | 3.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.3 | 1.1 | GO:0098989 | negative regulation of growth hormone secretion(GO:0060125) NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.3 | 6.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 1.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.3 | 4.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.3 | 6.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 2.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 2.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 13.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 1.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 0.7 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 6.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 1.8 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.2 | 5.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 5.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 7.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 7.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 3.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.0 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 3.7 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.2 | 9.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 3.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 | 3.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 3.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 2.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 2.1 | GO:0021684 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) prepulse inhibition(GO:0060134) |
| 0.2 | 10.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 12.0 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 4.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.9 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.1 | 10.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 1.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 3.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 3.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 2.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.5 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.1 | 0.8 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.1 | 1.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 2.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 4.5 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 3.6 | GO:0006900 | membrane budding(GO:0006900) |
| 0.1 | 2.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 5.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 4.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 2.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 2.4 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.1 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 1.6 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 3.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 4.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 4.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.8 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.1 | 4.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 3.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.1 | 2.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 1.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.4 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.9 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.0 | 2.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0007060 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) gene conversion(GO:0035822) |
| 0.0 | 4.5 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 1.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 7.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 17.5 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 1.1 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 2.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 2.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 32.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.0 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.4 | GO:0030149 | sphingolipid catabolic process(GO:0030149) |
| 0.0 | 1.9 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 23.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.6 | 7.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.5 | 32.6 | GO:0042627 | chylomicron(GO:0042627) |
| 2.1 | 12.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.8 | 9.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.8 | 7.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.8 | 10.7 | GO:0030478 | actin cap(GO:0030478) |
| 1.7 | 5.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 1.6 | 24.3 | GO:0032982 | myosin filament(GO:0032982) |
| 1.6 | 7.9 | GO:0043293 | apoptosome(GO:0043293) |
| 1.6 | 7.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.5 | 9.3 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.3 | 9.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 1.2 | 8.7 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.2 | 10.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 1.1 | 4.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 4.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.1 | 13.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 1.0 | 3.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 1.0 | 5.7 | GO:0033093 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 3.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.9 | 7.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.8 | 2.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.8 | 4.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.7 | 19.5 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 2.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.7 | 11.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.6 | 1.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.5 | 5.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.5 | 5.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 3.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 11.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 1.0 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.3 | 8.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 4.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 5.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.3 | 11.2 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 5.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.3 | 3.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 5.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 3.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 0.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 1.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 11.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.3 | 3.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 0.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 5.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 7.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 4.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 10.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 4.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 6.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 14.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 3.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 4.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.2 | 2.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 10.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 11.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 8.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 46.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 37.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 10.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 19.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 3.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 24.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 8.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 5.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 3.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 4.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 11.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 5.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 94.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 9.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 3.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 11.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 4.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 28.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 5.2 | 20.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 3.8 | 11.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 2.7 | 8.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 2.4 | 11.9 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 2.3 | 9.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.2 | 6.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.2 | 8.7 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 2.1 | 8.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.9 | 7.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.9 | 5.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.8 | 12.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.8 | 17.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.7 | 5.2 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 1.7 | 22.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 1.5 | 5.8 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 1.4 | 4.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.4 | 5.8 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 1.4 | 7.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 1.4 | 10.1 | GO:0043426 | MRF binding(GO:0043426) |
| 1.4 | 7.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.4 | 2.9 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 1.4 | 4.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.4 | 6.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.3 | 7.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.2 | 10.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.2 | 7.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 1.1 | 4.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.1 | 15.6 | GO:0031432 | titin binding(GO:0031432) |
| 1.1 | 3.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.0 | 9.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.0 | 3.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.0 | 3.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.9 | 20.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.9 | 6.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.9 | 6.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.9 | 2.6 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.9 | 7.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 3.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.8 | 5.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.8 | 3.8 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.8 | 3.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.7 | 7.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.7 | 3.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.6 | 7.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 7.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 4.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.6 | 5.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.6 | 3.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.6 | 8.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.6 | 7.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 7.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.6 | 6.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 2.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.5 | 2.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 4.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.5 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 3.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.5 | 1.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.5 | 13.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.5 | 1.9 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 0.5 | 7.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 5.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 3.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 8.7 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 3.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.4 | 15.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 3.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 5.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 2.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 2.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.4 | 17.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.4 | 2.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 2.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 7.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 12.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 2.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 8.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 1.9 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 1.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.4 | 4.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 5.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 8.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 2.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 6.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 4.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 6.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 6.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 2.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 5.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 3.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 3.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.3 | 3.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 19.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 9.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 3.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 1.6 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 0.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 12.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 3.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 2.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 3.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 27.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 5.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 5.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 23.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 10.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 4.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 3.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 2.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.2 | 18.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 2.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 10.5 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.1 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 4.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 9.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 3.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 13.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 8.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 3.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 5.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 34.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.7 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 0.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 7.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 4.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 10.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 4.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 10.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 35.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.4 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 3.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.6 | GO:0008134 | transcription factor binding(GO:0008134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 38.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.8 | 16.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.7 | 13.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 9.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 29.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.5 | 3.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 24.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.4 | 5.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 23.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 13.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 12.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 13.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 10.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 1.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 14.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 14.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 5.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 33.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 5.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 11.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 3.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 10.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 3.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 8.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 3.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 8.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 6.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 4.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 3.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 25.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 4.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 5.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 7.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 20.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.4 | 25.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.1 | 28.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.0 | 12.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.0 | 2.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.9 | 8.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.7 | 19.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.7 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 19.9 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.7 | 8.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 23.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.6 | 5.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.6 | 8.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 9.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.4 | 9.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 17.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 8.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 15.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 3.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 5.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 11.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 13.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 5.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 6.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.4 | 14.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 18.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 5.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 7.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 3.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 3.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 4.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 5.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 7.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 19.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 5.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 3.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 11.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 5.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 4.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 9.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 11.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 7.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 17.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.8 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 3.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 7.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 5.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.4 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |