Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
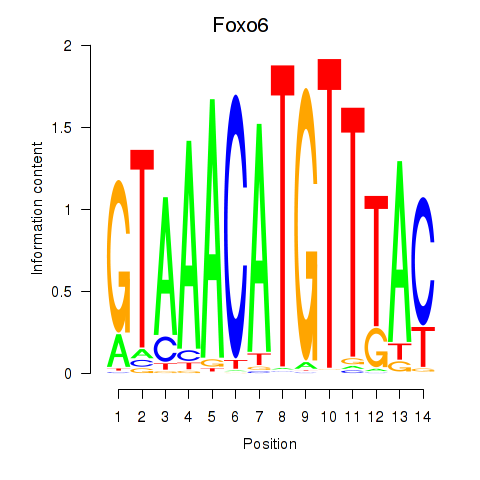
Results for Foxo6
Z-value: 0.48
Transcription factors associated with Foxo6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo6
|
ENSRNOG00000032639 | forkhead box O6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo6 | rn6_v1_chr5_-_139227196_139227196 | -0.21 | 1.2e-04 | Click! |
Activity profile of Foxo6 motif
Sorted Z-values of Foxo6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_49109981 | 20.37 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr2_+_93758919 | 12.98 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr14_+_22517774 | 12.85 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr14_-_19072677 | 12.19 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr5_-_54482159 | 11.01 |
ENSRNOT00000050030
|
RGD1306195
|
similar to RAN protein |
| chr18_-_15089988 | 10.86 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr5_-_33664435 | 8.72 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr9_+_77320726 | 8.44 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr2_+_248649441 | 8.28 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr11_+_52828116 | 8.28 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr2_+_23289374 | 8.27 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr15_+_80040842 | 8.04 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr3_+_143151739 | 7.69 |
ENSRNOT00000006850
|
Cst13
|
cystatin 13 |
| chr6_-_86223052 | 7.62 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr1_+_101474334 | 7.61 |
ENSRNOT00000064245
|
Tulp2
|
tubby-like protein 2 |
| chr1_-_224389389 | 7.60 |
ENSRNOT00000077408
ENSRNOT00000050010 |
UST4r
|
integral membrane transport protein UST4r |
| chr2_+_188543137 | 7.49 |
ENSRNOT00000027850
|
Muc1
|
mucin 1, cell surface associated |
| chr1_+_201687758 | 7.30 |
ENSRNOT00000093308
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_-_107114271 | 7.25 |
ENSRNOT00000004035
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr1_+_189359853 | 7.19 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr2_+_147006830 | 7.10 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chrX_+_45965301 | 6.95 |
ENSRNOT00000005141
|
Fam47a
|
family with sequence similarity 47, member A |
| chr5_-_173484986 | 6.83 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr4_+_120671489 | 6.76 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr1_-_253000760 | 6.62 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr3_-_156905892 | 6.47 |
ENSRNOT00000022424
|
Emilin3
|
elastin microfibril interfacer 3 |
| chr2_-_196415530 | 6.43 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr7_+_40316639 | 6.26 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr17_+_8558827 | 6.22 |
ENSRNOT00000016187
|
Il9
|
interleukin 9 |
| chr19_+_85606 | 6.22 |
ENSRNOT00000015724
|
Ces2e
|
carboxylesterase 2E |
| chr8_-_40883880 | 6.11 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chrX_+_15669191 | 6.10 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chr1_-_143398093 | 5.90 |
ENSRNOT00000078916
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chrX_+_25737292 | 5.66 |
ENSRNOT00000004893
|
RGD1563947
|
similar to RIKEN cDNA 4933400A11 |
| chr4_+_22084954 | 5.37 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr10_-_87564327 | 5.20 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr1_+_279896973 | 5.06 |
ENSRNOT00000068119
|
Pnliprp2
|
pancreatic lipase related protein 2 |
| chr5_+_64412895 | 4.97 |
ENSRNOT00000010561
|
Murc
|
muscle-related coiled-coil protein |
| chr9_-_117484262 | 4.96 |
ENSRNOT00000091931
|
AABR07068750.1
|
|
| chr4_+_153876149 | 4.91 |
ENSRNOT00000018083
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr16_+_68633720 | 4.87 |
ENSRNOT00000081838
|
LOC100911229
|
sperm motility kinase-like |
| chr20_+_915079 | 4.87 |
ENSRNOT00000074181
|
LOC100910382
|
olfactory receptor 14J1-like |
| chr1_-_171979933 | 4.81 |
ENSRNOT00000026744
|
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr18_+_29124428 | 4.72 |
ENSRNOT00000073131
|
Igip
|
IgA-inducing protein |
| chr11_+_88095170 | 4.71 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr17_+_59714657 | 4.70 |
ENSRNOT00000035675
|
RGD1560860
|
similar to ankyrin repeat domain 26 |
| chrX_-_37705263 | 4.59 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr1_-_102826965 | 4.48 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr1_+_125124743 | 4.43 |
ENSRNOT00000021530
|
Mtmr10
|
myotubularin related protein 10 |
| chrX_+_43183381 | 4.41 |
ENSRNOT00000039874
|
Magea8
|
melanoma antigen, family A, 8 |
| chr13_+_82552550 | 4.40 |
ENSRNOT00000003817
ENSRNOT00000076722 ENSRNOT00000076607 |
Slc19a2
|
solute carrier family 19 member 2 |
| chr2_-_228665279 | 4.36 |
ENSRNOT00000055648
|
RGD1306519
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr1_-_193328371 | 4.29 |
ENSRNOT00000019304
ENSRNOT00000031770 ENSRNOT00000019309 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chrX_-_143558521 | 4.25 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr1_+_177183209 | 4.22 |
ENSRNOT00000085326
|
Micalcl
|
MICAL C-terminal like |
| chr19_-_26006970 | 4.17 |
ENSRNOT00000004570
|
Gcdh
|
glutaryl-CoA dehydrogenase |
| chr10_-_41491554 | 3.90 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr5_-_160282810 | 3.79 |
ENSRNOT00000076160
ENSRNOT00000016352 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr10_-_90415070 | 3.72 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr17_+_38929045 | 3.59 |
ENSRNOT00000081739
|
Prl7d1
|
prolactin family 7, subfamily d, member 1 |
| chr17_-_42740021 | 3.45 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr10_+_13230158 | 3.42 |
ENSRNOT00000075648
|
Csap1
|
common salivary protein 1 |
| chr1_-_228558593 | 3.33 |
ENSRNOT00000028606
|
Olr326
|
olfactory receptor 326 |
| chr1_+_101012822 | 3.32 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr3_+_61756148 | 3.30 |
ENSRNOT00000002134
|
Mtx2
|
metaxin 2 |
| chr14_-_72122158 | 3.23 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_+_282694906 | 3.06 |
ENSRNOT00000074303
|
Ces2c
|
carboxylesterase 2C |
| chr18_-_399242 | 3.01 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr17_-_57765652 | 3.01 |
ENSRNOT00000044132
|
RGD1564999
|
similar to isopentenyl-diphosphate delta isomerase 2 |
| chr1_+_261464063 | 2.96 |
ENSRNOT00000082839
|
AABR07006871.1
|
|
| chr3_+_75299283 | 2.77 |
ENSRNOT00000013328
|
Olr555
|
olfactory receptor 555 |
| chr5_-_153737161 | 2.70 |
ENSRNOT00000073506
|
Ncmap
|
noncompact myelin associated protein |
| chr17_-_57394985 | 2.69 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr17_+_45670284 | 2.67 |
ENSRNOT00000086536
|
Olr1660
|
olfactory receptor 1660 |
| chr15_+_44329464 | 2.61 |
ENSRNOT00000044151
|
AABR07018164.1
|
|
| chr2_-_140464607 | 2.61 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr8_+_99636749 | 2.57 |
ENSRNOT00000086524
|
Plscr1
|
phospholipid scramblase 1 |
| chr15_+_46196771 | 2.53 |
ENSRNOT00000059212
|
Defb43
|
defensin beta 43 |
| chr20_+_29445510 | 2.51 |
ENSRNOT00000044805
|
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chrX_+_24059516 | 2.49 |
ENSRNOT00000049256
|
LOC685747
|
hypothetical protein LOC685747 |
| chrX_-_106085878 | 2.44 |
ENSRNOT00000049735
|
Prame
|
preferentially expressed antigen in melanoma |
| chr7_-_36408588 | 2.38 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_+_97559841 | 2.37 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr20_+_734642 | 2.30 |
ENSRNOT00000073543
|
Olr1688
|
olfactory receptor 1688 |
| chr3_+_72063504 | 2.26 |
ENSRNOT00000066606
|
Btbd18
|
BTB domain containing 18 |
| chr3_-_79121620 | 2.25 |
ENSRNOT00000087141
|
Olr741
|
olfactory receptor 741 |
| chr1_-_275876329 | 2.25 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_-_247088012 | 2.10 |
ENSRNOT00000020464
|
Spata6l
|
spermatogenesis associated 6-like |
| chr8_-_17860348 | 2.08 |
ENSRNOT00000046875
|
Olr1118
|
olfactory receptor 1118 |
| chr9_-_99659425 | 2.05 |
ENSRNOT00000051686
|
Olr1343
|
olfactory receptor 1343 |
| chr4_+_82637411 | 2.04 |
ENSRNOT00000048225
|
LOC685406
|
LRRGT00062 |
| chr3_-_121836086 | 2.01 |
ENSRNOT00000006113
|
Il1a
|
interleukin 1 alpha |
| chr3_-_74342719 | 1.94 |
ENSRNOT00000013060
|
Olr519
|
olfactory receptor 519 |
| chr16_-_49453394 | 1.93 |
ENSRNOT00000041617
|
Lrp2bp
|
Lrp2 binding protein |
| chr10_+_111716966 | 1.91 |
ENSRNOT00000070942
|
LOC100911926
|
vomeronasal type-2 receptor 26-like |
| chr2_-_206842724 | 1.83 |
ENSRNOT00000086252
|
AABR07012763.1
|
|
| chr5_+_12406988 | 1.82 |
ENSRNOT00000061899
|
RGD1564095
|
similar to 60S acidic ribosomal protein P2 |
| chr3_+_123776181 | 1.81 |
ENSRNOT00000090764
ENSRNOT00000034146 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr7_+_120465130 | 1.73 |
ENSRNOT00000016077
|
Pick1
|
protein interacting with PRKCA 1 |
| chr14_+_7949239 | 1.70 |
ENSRNOT00000044617
|
Ndufb4l1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4-like 1 |
| chr8_+_42284654 | 1.64 |
ENSRNOT00000043810
|
LOC100911939
|
olfactory receptor 8B3-like |
| chr3_-_76114902 | 1.59 |
ENSRNOT00000071564
|
LOC102555599
|
olfactory receptor 5L1-like |
| chr13_-_45040593 | 1.56 |
ENSRNOT00000004908
|
Lct
|
lactase |
| chr7_-_139784444 | 1.56 |
ENSRNOT00000084656
|
AC110306.1
|
|
| chr1_-_149441437 | 1.53 |
ENSRNOT00000041780
|
Vom2r41
|
vomeronasal 2 receptor, 41 |
| chr14_-_21615488 | 1.51 |
ENSRNOT00000002667
|
Vcsa2
|
variable coding sequence A2 |
| chr17_+_3210130 | 1.51 |
ENSRNOT00000072334
|
RGD1564657
|
similar to cathepsin 1 precursor |
| chr6_-_26138414 | 1.45 |
ENSRNOT00000034712
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chr4_+_62019970 | 1.44 |
ENSRNOT00000013133
|
LOC100910708
|
aldose reductase-related protein 1-like |
| chr4_-_1752548 | 1.43 |
ENSRNOT00000071677
|
LOC100912415
|
olfactory receptor 150-like |
| chr10_-_6870011 | 1.40 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr7_+_13631975 | 1.38 |
ENSRNOT00000083548
|
Olr1084
|
olfactory receptor 1084 |
| chr3_-_78717401 | 1.35 |
ENSRNOT00000008676
|
Olr720
|
olfactory receptor 720 |
| chr2_+_115092263 | 1.30 |
ENSRNOT00000064455
|
LOC100910543
|
olfactory receptor 150-like |
| chr8_-_3882077 | 1.29 |
ENSRNOT00000071085
|
Vom2r25
|
vomeronasal 2 receptor, 25 |
| chr15_+_35571103 | 1.28 |
ENSRNOT00000085376
|
Olr1293
|
olfactory receptor 1293 |
| chr9_-_17120677 | 1.27 |
ENSRNOT00000025769
|
Yipf3
|
Yip1 domain family, member 3 |
| chr1_-_63571794 | 1.25 |
ENSRNOT00000046049
|
Vom2r30
|
vomeronasal 2 receptor, 30 |
| chr2_-_127781003 | 1.24 |
ENSRNOT00000050764
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr1_-_242152834 | 1.24 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr4_+_45854403 | 1.23 |
ENSRNOT00000084484
|
LOC685171
|
similar to protein disulfide isomerase-associated 6 |
| chr4_-_167180656 | 1.16 |
ENSRNOT00000049487
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr19_+_23832780 | 1.10 |
ENSRNOT00000045060
|
AABR07043141.1
|
|
| chr6_-_141715846 | 1.07 |
ENSRNOT00000044966
|
AABR07065798.1
|
|
| chr1_-_150101177 | 1.02 |
ENSRNOT00000042931
|
Olr24
|
olfactory receptor 24 |
| chr10_+_87782376 | 0.93 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chr9_+_17120759 | 0.88 |
ENSRNOT00000025787
|
Polr1c
|
RNA polymerase I subunit C |
| chr1_+_60044985 | 0.88 |
ENSRNOT00000079924
|
Vom1r6
|
vomeronasal 1 receptor 6 |
| chr8_+_44638556 | 0.87 |
ENSRNOT00000044669
|
Vom2r3
|
vomeronasal 2 receptor, 3 |
| chr1_-_149175205 | 0.85 |
ENSRNOT00000089091
ENSRNOT00000045611 ENSRNOT00000089015 |
Vom2r42
|
vomeronasal 2 receptor, 42 |
| chr7_+_6096690 | 0.84 |
ENSRNOT00000051694
|
Olr1006
|
olfactory receptor 1006 |
| chr5_+_116973159 | 0.83 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr3_+_75848904 | 0.83 |
ENSRNOT00000042996
|
Olr583
|
olfactory receptor 583 |
| chr13_+_96195836 | 0.82 |
ENSRNOT00000042547
|
RGD1563812
|
similar to basic transcription factor 3 |
| chr14_+_779169 | 0.82 |
ENSRNOT00000052336
ENSRNOT00000073211 ENSRNOT00000086687 |
AABR07014087.1
|
|
| chr3_+_73988945 | 0.81 |
ENSRNOT00000050404
|
Olr515
|
olfactory receptor 515 |
| chr3_+_15823220 | 0.81 |
ENSRNOT00000075496
|
Olr398
|
olfactory receptor 398 |
| chr7_+_9294709 | 0.78 |
ENSRNOT00000048396
|
Olr1065
|
olfactory receptor 1065 |
| chr1_-_172215730 | 0.74 |
ENSRNOT00000055175
|
Olr244
|
olfactory receptor 244 |
| chr19_-_37216572 | 0.74 |
ENSRNOT00000020405
|
Tradd
|
TNFRSF1A-associated via death domain |
| chr15_+_18941431 | 0.74 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr16_-_75028977 | 0.73 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr15_-_27408258 | 0.71 |
ENSRNOT00000043588
|
RGD1562558
|
similar to Cytochrome c, somatic |
| chr1_-_69016426 | 0.68 |
ENSRNOT00000093436
|
Vom2r3
|
|
| chr9_+_44491177 | 0.67 |
ENSRNOT00000075242
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr1_-_171045419 | 0.63 |
ENSRNOT00000047239
|
Olr224
|
olfactory receptor 224 |
| chr20_+_8484311 | 0.61 |
ENSRNOT00000050041
|
LOC100364116
|
ribosomal protein S20-like |
| chr4_+_121760773 | 0.60 |
ENSRNOT00000087069
|
Vom1r92
|
vomeronasal 1 receptor 92 |
| chr3_+_76179214 | 0.56 |
ENSRNOT00000071281
|
LOC686677
|
similar to olfactory receptor 1161 |
| chr3_-_73839527 | 0.55 |
ENSRNOT00000080756
|
Olr508
|
olfactory receptor 508 |
| chr12_-_47987255 | 0.54 |
ENSRNOT00000074000
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr1_+_170796271 | 0.50 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr14_+_22192970 | 0.50 |
ENSRNOT00000041514
ENSRNOT00000002704 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr10_-_60573129 | 0.48 |
ENSRNOT00000083428
|
Olr1499
|
olfactory receptor 1499 |
| chr3_+_19071980 | 0.47 |
ENSRNOT00000079487
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chrX_-_75224268 | 0.43 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr4_-_103258134 | 0.42 |
ENSRNOT00000086827
|
AABR07061052.1
|
|
| chr7_-_116106368 | 0.41 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr11_-_65350442 | 0.40 |
ENSRNOT00000003773
|
Gpr156
|
G protein-coupled receptor 156 |
| chrX_-_156999650 | 0.40 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr17_+_70586394 | 0.37 |
ENSRNOT00000025550
|
Rbm17
|
RNA binding motif protein 17 |
| chr1_-_60332899 | 0.34 |
ENSRNOT00000078636
|
LOC108348087
|
vomeronasal type-1 receptor 4-like |
| chr8_+_41358307 | 0.32 |
ENSRNOT00000071307
|
LOC100911801
|
olfactory receptor 143-like |
| chr8_+_5676665 | 0.32 |
ENSRNOT00000012310
|
Mmp3
|
matrix metallopeptidase 3 |
| chr4_+_122124451 | 0.31 |
ENSRNOT00000052101
|
V1ra14
|
vomeronasal 1 receptor A14 |
| chr16_-_20641908 | 0.30 |
ENSRNOT00000026846
|
Ell
|
elongation factor for RNA polymerase II |
| chr11_+_42945084 | 0.30 |
ENSRNOT00000002292
|
Crybg3
|
crystallin beta-gamma domain containing 3 |
| chr3_-_12029877 | 0.30 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr7_+_9262982 | 0.29 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr1_-_73961432 | 0.27 |
ENSRNOT00000035435
|
Nlrp4b
|
NLR family, pyrin domain containing 4B |
| chr4_-_167068838 | 0.26 |
ENSRNOT00000029713
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chr15_+_75067297 | 0.25 |
ENSRNOT00000057931
|
LOC498555
|
similar to 60S acidic ribosomal protein P2 |
| chr15_+_28413461 | 0.23 |
ENSRNOT00000060521
|
Tmem253
|
transmembrane protein 253 |
| chr13_-_35933243 | 0.22 |
ENSRNOT00000039074
|
Tmem177
|
transmembrane protein 177 |
| chr7_-_17859653 | 0.22 |
ENSRNOT00000030164
|
Zscan4f
|
zinc finger and SCAN domain containing 4F |
| chr3_+_15688992 | 0.21 |
ENSRNOT00000045037
|
Olr395
|
olfactory receptor 395 |
| chr3_+_19366370 | 0.21 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr1_-_170916059 | 0.20 |
ENSRNOT00000041974
|
Olr217
|
olfactory receptor 217 |
| chr12_+_41200718 | 0.20 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr3_-_113057791 | 0.18 |
ENSRNOT00000056190
|
Tgm7l1
|
transglutaminase 7-like 1 |
| chr9_+_101222846 | 0.16 |
ENSRNOT00000074328
|
Vom1r64
|
vomeronasal 1 receptor 64 |
| chr4_+_87440996 | 0.15 |
ENSRNOT00000074184
|
Vom1r68
|
vomeronasal 1 receptor 68 |
| chr1_-_74638587 | 0.15 |
ENSRNOT00000046047
|
Vom2r30
|
vomeronasal 2 receptor, 30 |
| chr4_+_166993669 | 0.09 |
ENSRNOT00000045376
|
Tas2r109
|
taste receptor, type 2, member 109 |
| chr2_+_87957363 | 0.06 |
ENSRNOT00000047881
|
LOC499565
|
LRRGT00038 |
| chr4_+_25635765 | 0.05 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr1_-_193700673 | 0.04 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr18_-_71614980 | 0.03 |
ENSRNOT00000032563
|
LOC102548286
|
peroxisomal biogenesis factor 19-like |
| chr19_+_25946979 | 0.02 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr1_-_63457134 | 0.01 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr18_+_61377051 | 0.01 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr1_-_60281386 | 0.00 |
ENSRNOT00000092184
|
Vom1r10
|
vomeronasal 1 receptor 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.8 | 8.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.3 | 6.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 1.7 | 8.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.5 | 4.4 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 1.3 | 7.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 1.2 | 4.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.1 | 5.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.9 | 7.5 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.9 | 2.7 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.8 | 3.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.7 | 5.1 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 0.7 | 7.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.7 | 2.0 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 0.6 | 10.8 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.5 | 1.8 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.4 | 4.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 1.2 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.4 | 6.2 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.4 | 2.6 | GO:0070782 | phosphatidylserine biosynthetic process(GO:0006659) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.4 | 7.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 1.7 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 | 2.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 1.6 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.3 | 2.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 4.9 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 10.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 6.6 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.2 | 4.9 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 2.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 2.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 3.6 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 4.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 7.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 4.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.7 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 3.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0071460 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 2.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 1.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 27.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 3.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 4.8 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 2.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.7 | 12.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 7.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 6.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 4.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 4.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 7.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 2.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 3.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 6.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 7.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 6.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 8.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 4.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 34.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 7.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.1 | 8.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 1.8 | 5.4 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 1.5 | 4.4 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 1.0 | 7.3 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.9 | 7.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 4.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.8 | 4.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.8 | 3.1 | GO:0047619 | acylcarnitine hydrolase activity(GO:0047619) |
| 0.8 | 7.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 9.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 3.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.5 | 1.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.5 | 8.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.4 | 13.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 6.6 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.4 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 4.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 3.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 4.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 2.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 6.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 4.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 7.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 7.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 9.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 3.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 20.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 8.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 8.7 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 3.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 7.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 8.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 7.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 13.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 1.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 5.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 1.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 3.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 5.1 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.1 | 7.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 4.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 5.5 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 4.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |