Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
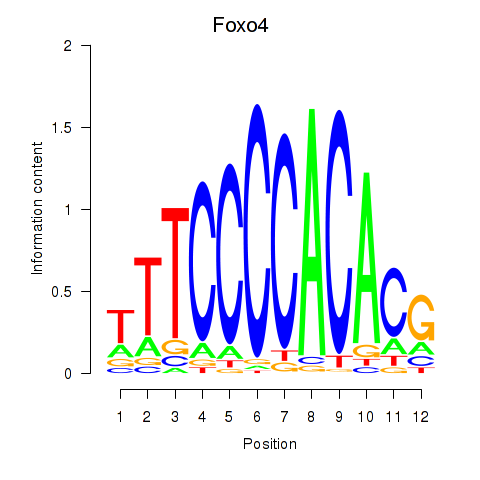
Results for Foxo4
Z-value: 0.55
Transcription factors associated with Foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo4
|
ENSRNOG00000033316 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo4 | rn6_v1_chrX_+_71155894_71155894 | 0.02 | 6.6e-01 | Click! |
Activity profile of Foxo4 motif
Sorted Z-values of Foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_168957460 | 31.45 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr1_+_168945449 | 31.37 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr1_-_168972725 | 30.29 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr4_-_70996395 | 20.10 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr3_+_16817051 | 12.64 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr9_+_67774150 | 10.41 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr5_-_139921013 | 9.76 |
ENSRNOT00000079957
|
Smap2
|
small ArfGAP2 |
| chr1_-_216828581 | 9.56 |
ENSRNOT00000066943
ENSRNOT00000088856 |
Tnfrsf26
|
tumor necrosis factor receptor superfamily, member 26 |
| chr15_-_51834030 | 6.19 |
ENSRNOT00000024895
|
Ccar2
|
cell cycle and apoptosis regulator 2 |
| chr17_+_76532611 | 6.14 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chrX_+_71342775 | 6.03 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr1_+_213737788 | 5.57 |
ENSRNOT00000020004
|
Pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr20_-_157861 | 5.45 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr5_-_139933764 | 5.34 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr20_+_5414448 | 4.86 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chrX_+_112555173 | 4.63 |
ENSRNOT00000045023
|
LOC103690888
|
60S ribosomal protein L10a-like |
| chr8_+_40009691 | 4.50 |
ENSRNOT00000042679
|
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr5_+_148577332 | 4.49 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr14_+_88549947 | 4.44 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr1_+_226252226 | 4.39 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr10_-_18090932 | 4.20 |
ENSRNOT00000064744
ENSRNOT00000006591 |
Npm1
|
nucleophosmin 1 |
| chr19_+_9436210 | 4.13 |
ENSRNOT00000074845
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr14_+_86652365 | 4.08 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr6_-_135304422 | 4.00 |
ENSRNOT00000010598
|
Cinp
|
cyclin-dependent kinase 2-interacting protein |
| chr7_-_92882068 | 3.94 |
ENSRNOT00000037809
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr3_+_91870208 | 3.58 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr7_-_71139267 | 3.37 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr9_+_42620006 | 3.21 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr3_-_138116664 | 3.17 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chr1_-_84168044 | 3.02 |
ENSRNOT00000084586
ENSRNOT00000028348 |
Shkbp1
|
Sh3kbp1 binding protein 1 |
| chr3_+_2877293 | 2.88 |
ENSRNOT00000061855
|
Lcn5
|
lipocalin 5 |
| chr1_-_80483487 | 2.82 |
ENSRNOT00000074418
|
Gemin7
|
gem (nuclear organelle) associated protein 7 |
| chr3_+_149144795 | 2.49 |
ENSRNOT00000015482
|
Dnmt3b
|
DNA methyltransferase 3 beta |
| chr14_-_1632378 | 2.36 |
ENSRNOT00000000080
|
Dr1
|
down-regulator of transcription 1 |
| chr3_-_13824870 | 2.34 |
ENSRNOT00000066961
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr6_+_126623015 | 2.26 |
ENSRNOT00000010672
|
Tmem251
|
transmembrane protein 251 |
| chr1_-_199320727 | 2.15 |
ENSRNOT00000022452
|
Zfp668
|
zinc finger protein 668 |
| chr5_-_154363329 | 2.14 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chr14_+_83510278 | 2.10 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_+_83510640 | 2.04 |
ENSRNOT00000089928
ENSRNOT00000082242 ENSRNOT00000025378 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr5_-_148577292 | 1.98 |
ENSRNOT00000017156
|
Zcchc17
|
zinc finger CCHC-type containing 17 |
| chr5_-_48194251 | 1.79 |
ENSRNOT00000081748
ENSRNOT00000065912 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr10_+_85744568 | 1.75 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chr9_+_112360419 | 1.65 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr20_-_28263037 | 1.65 |
ENSRNOT00000030348
|
Edar
|
ectodysplasin-A receptor |
| chr1_-_82120902 | 1.54 |
ENSRNOT00000027684
|
Erf
|
Ets2 repressor factor |
| chr10_-_90049112 | 1.32 |
ENSRNOT00000028323
|
Pyy
|
peptide YY (mapped) |
| chr4_+_159622404 | 1.29 |
ENSRNOT00000078299
|
Fgf23
|
fibroblast growth factor 23 |
| chr10_+_97212432 | 1.13 |
ENSRNOT00000088599
|
Axin2
|
axin 2 |
| chr7_-_144223429 | 1.08 |
ENSRNOT00000020527
|
Atf7
|
activating transcription factor 7 |
| chr18_+_66741144 | 0.91 |
ENSRNOT00000021161
|
AABR07032457.1
|
|
| chr9_+_16427589 | 0.90 |
ENSRNOT00000091809
|
Gltscr1l
|
GLTSCR1-like |
| chr20_+_26534535 | 0.69 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr1_-_82236179 | 0.59 |
ENSRNOT00000027845
|
Cnfn
|
cornifelin |
| chr6_+_48452369 | 0.44 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr19_+_46733633 | 0.16 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chr4_-_66624912 | 0.12 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr3_-_74991168 | 0.11 |
ENSRNOT00000049273
|
Olr550
|
olfactory receptor 550 |
| chr10_+_93811350 | 0.03 |
ENSRNOT00000077280
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 30.3 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 4.5 | 62.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 3.4 | 20.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.6 | 4.9 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 1.4 | 4.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 1.3 | 3.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.1 | 3.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.0 | 6.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.8 | 2.5 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.7 | 4.4 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.6 | 6.2 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.6 | 10.4 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.4 | 1.3 | GO:1904383 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) response to sodium phosphate(GO:1904383) |
| 0.4 | 1.1 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of centromeric sister chromatid cohesion(GO:0070602) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 1.7 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 1.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 4.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 1.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 1.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 5.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 9.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.3 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.1 | 2.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 6.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.1 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 12.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 4.1 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 10.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 93.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.1 | 6.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 1.5 | 4.4 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.6 | 10.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 4.2 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 2.8 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 2.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 4.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 10.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 7.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 6.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 10.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 30.3 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 6.3 | 62.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.7 | 9.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 4.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.6 | 4.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.5 | 2.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 4.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 5.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.3 | 1.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 6.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 3.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 6.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 20.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 5.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.7 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 3.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 4.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 6.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 12.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 9.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 30.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 10.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 4.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 4.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 4.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 3.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |