Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
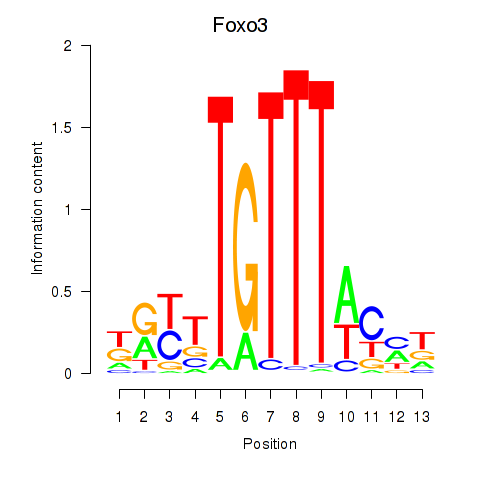
Results for Foxo3
Z-value: 0.66
Transcription factors associated with Foxo3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo3
|
ENSRNOG00000000299 | forkhead box O3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo3 | rn6_v1_chr20_+_46429222_46429222 | 0.07 | 2.1e-01 | Click! |
Activity profile of Foxo3 motif
Sorted Z-values of Foxo3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_64818813 | 27.88 |
ENSRNOT00000009111
ENSRNOT00000086505 |
Aldob
|
aldolase, fructose-bisphosphate B |
| chr7_-_119689938 | 20.71 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr13_+_91080341 | 19.42 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr4_+_154391647 | 18.53 |
ENSRNOT00000081488
ENSRNOT00000079192 |
Mug1
|
murinoglobulin 1 |
| chr14_+_87448692 | 17.73 |
ENSRNOT00000077177
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr20_-_54517709 | 17.49 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr13_+_89597138 | 16.51 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr2_+_262914327 | 15.69 |
ENSRNOT00000029312
|
Negr1
|
neuronal growth regulator 1 |
| chr1_+_234252757 | 15.15 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr1_+_88750462 | 15.13 |
ENSRNOT00000028247
|
LOC108348122
|
CAP-Gly domain-containing linker protein 3 |
| chr3_-_937102 | 14.28 |
ENSRNOT00000007471
|
Hnmt
|
histamine N-methyltransferase |
| chr16_+_50049828 | 13.48 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr10_+_86399827 | 13.01 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr1_-_90978907 | 12.54 |
ENSRNOT00000072819
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr10_+_89285855 | 12.18 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr10_+_89286047 | 10.98 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr5_+_124690214 | 10.76 |
ENSRNOT00000011237
|
Plpp3
|
phospholipid phosphatase 3 |
| chr2_-_258997138 | 9.22 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chrX_+_65040775 | 8.79 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr6_-_27487284 | 8.71 |
ENSRNOT00000084344
|
Selenoi
|
selenoprotein I |
| chrX_+_65040934 | 8.40 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr3_+_131351587 | 8.16 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr10_-_25845634 | 7.04 |
ENSRNOT00000004269
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr1_-_131454689 | 6.91 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_61658245 | 6.63 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr10_-_46720907 | 6.56 |
ENSRNOT00000083093
ENSRNOT00000067866 |
Tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr6_-_44363915 | 6.50 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr10_-_71849293 | 6.49 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr14_-_44845218 | 5.52 |
ENSRNOT00000004003
|
Klhl5
|
kelch-like family member 5 |
| chr2_-_188596222 | 5.41 |
ENSRNOT00000027920
|
Efna1
|
ephrin A1 |
| chr18_+_44810388 | 5.38 |
ENSRNOT00000021646
ENSRNOT00000089750 |
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr15_-_28313841 | 5.24 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr10_-_81942188 | 5.01 |
ENSRNOT00000003784
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr11_-_15858281 | 4.58 |
ENSRNOT00000090586
|
AABR07033287.1
|
|
| chr14_+_104191517 | 4.48 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr6_-_51018050 | 4.13 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr18_+_13386133 | 4.07 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chr15_+_2374582 | 3.99 |
ENSRNOT00000019644
|
Zfp503
|
zinc finger protein 503 |
| chr7_-_107775712 | 3.70 |
ENSRNOT00000010811
ENSRNOT00000093459 |
Ndrg1
|
N-myc downstream regulated 1 |
| chr3_+_116899878 | 3.68 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr3_-_37480984 | 3.58 |
ENSRNOT00000030373
|
Nmi
|
N-myc (and STAT) interactor |
| chr2_-_30634243 | 3.33 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr7_-_120770435 | 3.32 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr4_+_41364441 | 3.02 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr15_-_108120279 | 2.98 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr14_-_8432195 | 2.80 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_+_85417838 | 2.73 |
ENSRNOT00000012257
|
Ick
|
intestinal cell kinase |
| chr11_+_73693814 | 2.41 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr3_-_21148897 | 2.22 |
ENSRNOT00000010501
|
Olr425
|
olfactory receptor 425 |
| chr7_+_59326518 | 2.22 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr18_-_5314511 | 2.14 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr19_+_38601786 | 2.04 |
ENSRNOT00000087655
|
Zfp90
|
zinc finger protein 90 |
| chr3_+_80614937 | 1.94 |
ENSRNOT00000065462
|
Harbi1
|
harbinger transposase derived 1 |
| chr2_-_98610368 | 1.91 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr13_-_30800451 | 1.87 |
ENSRNOT00000046791
|
Rpl21
|
ribosomal protein L21 |
| chr2_+_184243976 | 1.76 |
ENSRNOT00000042146
|
Dear
|
dual endothelin 1, angiotensin II receptor |
| chr18_+_3887419 | 1.61 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr4_-_115516296 | 1.59 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chrX_+_10430847 | 1.50 |
ENSRNOT00000047936
|
Rpl21
|
ribosomal protein L21 |
| chr3_+_91870208 | 1.49 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr20_+_46519431 | 1.43 |
ENSRNOT00000077765
|
AABR07045405.1
|
|
| chr3_-_58009514 | 1.40 |
ENSRNOT00000048356
|
AABR07052498.1
|
|
| chr2_-_34254968 | 1.32 |
ENSRNOT00000016439
|
Trappc13
|
trafficking protein particle complex 13 |
| chr2_+_34255305 | 1.28 |
ENSRNOT00000016649
ENSRNOT00000016596 |
Trim23
|
tripartite motif-containing 23 |
| chr17_-_74679696 | 1.16 |
ENSRNOT00000048420
|
AABR07028558.1
|
|
| chr13_-_91638661 | 1.04 |
ENSRNOT00000045429
|
Olr1579
|
olfactory receptor 1579 |
| chr10_+_11393103 | 1.01 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr1_-_170801560 | 0.97 |
ENSRNOT00000047852
|
Olr211
|
olfactory receptor 211 |
| chr9_+_42315682 | 0.95 |
ENSRNOT00000071995
|
AABR07067388.1
|
|
| chr8_+_32604365 | 0.89 |
ENSRNOT00000071210
|
AABR07069587.1
|
|
| chr17_-_84705052 | 0.78 |
ENSRNOT00000050803
|
AABR07028749.1
|
|
| chr10_-_87335823 | 0.74 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr16_-_69280109 | 0.69 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr4_+_152630469 | 0.54 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr10_-_72556564 | 0.52 |
ENSRNOT00000048373
|
AABR07030162.1
|
|
| chr9_-_66627007 | 0.51 |
ENSRNOT00000049571
|
AABR07067829.1
|
|
| chr20_+_11649814 | 0.43 |
ENSRNOT00000073516
|
LOC103694398
|
keratin-associated protein 10-1-like |
| chr16_+_27399467 | 0.35 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr1_-_173393390 | 0.29 |
ENSRNOT00000050657
|
Olr285
|
olfactory receptor 285 |
| chr1_+_190671696 | 0.28 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr6_+_42947334 | 0.24 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr3_+_63536166 | 0.20 |
ENSRNOT00000016132
|
Plekha3
|
pleckstrin homology domain containing A3 |
| chr9_-_121972055 | 0.18 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chrX_-_20359945 | 0.11 |
ENSRNOT00000049138
|
AABR07037412.1
|
|
| chr16_-_32299542 | 0.10 |
ENSRNOT00000072321
|
AABR07025299.1
|
|
| chr18_+_47740328 | 0.01 |
ENSRNOT00000025119
|
Sncaip
|
synuclein, alpha interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 27.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 5.5 | 16.5 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 4.0 | 4.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 3.6 | 10.8 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 3.6 | 14.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 3.5 | 27.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 3.2 | 19.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 2.3 | 23.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 2.3 | 6.9 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 2.2 | 6.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.9 | 20.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.8 | 5.4 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 1.7 | 6.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.6 | 6.5 | GO:0060066 | metanephric part of ureteric bud development(GO:0035502) oviduct development(GO:0060066) |
| 1.4 | 7.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.4 | 5.4 | GO:0014028 | notochord formation(GO:0014028) |
| 1.3 | 5.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.3 | 15.2 | GO:0046548 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) retinal cone cell development(GO:0046549) |
| 1.0 | 13.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.9 | 3.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.9 | 17.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.7 | 2.7 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.6 | 17.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 8.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 3.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 | 1.8 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.3 | 3.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 18.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 2.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 4.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 3.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 6.6 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 3.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 7.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 15.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 8.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 3.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 8.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.0 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 2.1 | GO:0048663 | neuron fate commitment(GO:0048663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 27.9 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 2.9 | 17.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.3 | 7.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.8 | 16.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.0 | 27.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 3.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 1.6 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 13.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 21.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 23.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 19.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 6.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 18.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 8.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 5.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 5.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 4.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 5.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 36.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 22.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 6.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.3 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 4.8 | 14.3 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 4.6 | 23.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 4.0 | 27.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 4.0 | 27.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 3.6 | 10.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 3.0 | 15.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.9 | 8.7 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 2.8 | 19.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.5 | 17.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.0 | 17.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.8 | 5.4 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.6 | 4.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 6.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.8 | GO:0004962 | angiotensin type II receptor activity(GO:0004945) endothelin receptor activity(GO:0004962) |
| 0.3 | 3.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 23.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 20.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 6.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 3.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 9.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 6.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 13.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 6.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 8.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.4 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 68.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 5.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 13.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 19.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 14.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 6.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.1 | 17.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.0 | 16.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 17.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 27.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 5.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 11.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 23.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 7.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 15.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 4.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 3.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |