Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
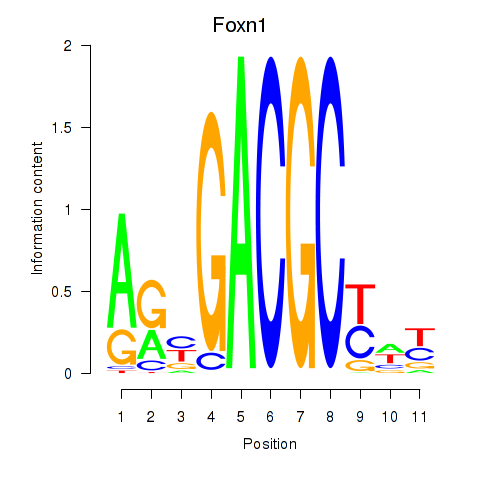
Results for Foxn1
Z-value: 0.27
Transcription factors associated with Foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxn1
|
ENSRNOG00000010870 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxn1 | rn6_v1_chr10_-_65634666_65634666 | 0.09 | 9.3e-02 | Click! |
Activity profile of Foxn1 motif
Sorted Z-values of Foxn1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_36760742 | 9.28 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr7_-_70552897 | 8.56 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr1_-_143647124 | 5.18 |
ENSRNOT00000026595
|
Btbd1
|
BTB domain containing 1 |
| chr9_-_114619711 | 5.00 |
ENSRNOT00000050012
|
Mtcl1
|
microtubule crosslinking factor 1 |
| chr2_+_4195917 | 4.35 |
ENSRNOT00000093303
|
RGD1560883
|
similar to KIAA0825 protein |
| chr6_+_109939345 | 3.77 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr2_-_4195810 | 3.76 |
ENSRNOT00000018457
|
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr13_+_96303703 | 3.75 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr18_+_55797198 | 3.66 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr20_-_6500523 | 3.52 |
ENSRNOT00000000629
|
Cpne5
|
copine 5 |
| chr1_-_255557055 | 2.84 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr1_-_165382216 | 2.81 |
ENSRNOT00000023648
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr1_-_220165678 | 2.56 |
ENSRNOT00000026915
|
Bbs1
|
Bardet-Biedl syndrome 1 |
| chr5_-_156811650 | 2.31 |
ENSRNOT00000068065
|
Fam43b
|
family with sequence similarity 43, member B |
| chr5_+_140538260 | 2.31 |
ENSRNOT00000017998
|
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr7_+_123102493 | 2.18 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chr10_+_85301875 | 2.14 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr8_-_50277797 | 2.13 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr8_-_70215719 | 2.01 |
ENSRNOT00000015598
|
Rab11a
|
RAB11a, member RAS oncogene family |
| chr13_-_79801112 | 1.93 |
ENSRNOT00000087323
ENSRNOT00000036483 |
Suco
|
SUN domain containing ossification factor |
| chr1_+_265904566 | 1.73 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr13_-_79801368 | 1.69 |
ENSRNOT00000075998
ENSRNOT00000084058 |
Suco
|
SUN domain containing ossification factor |
| chr16_-_20486707 | 1.61 |
ENSRNOT00000026470
|
Jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_58195143 | 1.59 |
ENSRNOT00000033706
|
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr8_-_114892910 | 1.39 |
ENSRNOT00000074934
|
Ppm1m
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr10_-_56850085 | 1.38 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr13_-_79801561 | 1.34 |
ENSRNOT00000075936
|
Suco
|
SUN domain containing ossification factor |
| chr6_+_10533151 | 1.18 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr10_-_88645364 | 0.74 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr12_+_47074200 | 0.73 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr9_+_102862890 | 0.64 |
ENSRNOT00000050494
ENSRNOT00000080129 |
Fam174a
|
family with sequence similarity 174, member A |
| chr19_-_24831500 | 0.52 |
ENSRNOT00000005770
|
Pkn1
|
protein kinase N1 |
| chr3_+_11424099 | 0.48 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chr19_-_26094756 | 0.42 |
ENSRNOT00000067780
|
Junb
|
JunB proto-oncogene, AP-1 transcription factor subunit |
| chr14_+_82093837 | 0.35 |
ENSRNOT00000021593
|
Nelfa
|
negative elongation factor complex member A |
| chr3_+_101156212 | 0.19 |
ENSRNOT00000065333
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr5_+_156810991 | 0.13 |
ENSRNOT00000055904
|
AABR07050222.1
|
|
| chr13_-_26768708 | 0.10 |
ENSRNOT00000092763
|
Bcl2
|
BCL2, apoptosis regulator |
| chr6_-_2729394 | 0.07 |
ENSRNOT00000009175
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxn1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.3 | 3.8 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 1.0 | 5.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.9 | 2.6 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.7 | 2.0 | GO:0036258 | regulation of multivesicular body size(GO:0010796) multivesicular body assembly(GO:0036258) |
| 0.6 | 1.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.6 | 2.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.4 | 4.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.4 | 2.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.4 | 2.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 3.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 2.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 9.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 5.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 3.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 2.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.5 | GO:0002634 | regulation of germinal center formation(GO:0002634) histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 1.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 5.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.6 | 8.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 3.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 9.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 3.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 2.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 3.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 2.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 2.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 5.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 5.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.7 | 2.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.5 | 8.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 2.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 2.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 9.3 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 2.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 3.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 5.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 4.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |