Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
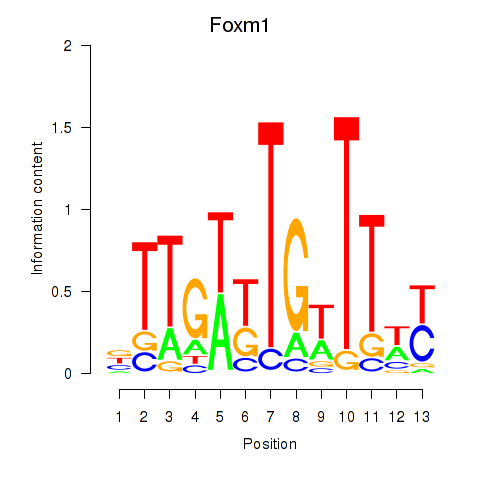
Results for Foxm1
Z-value: 0.59
Transcription factors associated with Foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxm1
|
ENSRNOG00000005936 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxm1 | rn6_v1_chr4_+_161685258_161685283 | -0.19 | 6.1e-04 | Click! |
Activity profile of Foxm1 motif
Sorted Z-values of Foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_3677474 | 20.87 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr3_+_51883559 | 14.36 |
ENSRNOT00000007197
|
Csrnp3
|
cysteine and serine rich nuclear protein 3 |
| chr17_+_78793336 | 12.49 |
ENSRNOT00000057898
|
Mt1
|
metallothionein 1 |
| chr18_-_26211445 | 12.12 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr10_+_86399827 | 9.67 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr16_+_72388880 | 9.45 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr16_+_72401887 | 9.35 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr1_+_107262659 | 8.93 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr8_-_115981910 | 7.84 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr10_+_59894340 | 7.77 |
ENSRNOT00000080446
|
Spata22
|
spermatogenesis associated 22 |
| chr16_+_18736154 | 7.46 |
ENSRNOT00000015723
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr7_-_101138860 | 7.40 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr18_-_61788859 | 7.15 |
ENSRNOT00000034075
ENSRNOT00000034069 |
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr2_+_182006242 | 7.03 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr14_+_12218553 | 6.90 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr16_+_74531564 | 6.58 |
ENSRNOT00000078971
|
Slc25a15
|
solute carrier family 25 member 15 |
| chr9_+_112360419 | 6.17 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr7_-_101138373 | 6.05 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr8_+_116332796 | 5.84 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr7_+_121408829 | 5.51 |
ENSRNOT00000023434
|
Mgat3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr1_-_236900904 | 5.32 |
ENSRNOT00000066846
|
AABR07006480.1
|
|
| chr11_-_87924816 | 5.32 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr1_+_258074860 | 5.10 |
ENSRNOT00000054729
|
Cyp2c24
|
cytochrome P450, family 2, subfamily c, polypeptide 24 |
| chr18_-_58423196 | 5.02 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr1_+_147713892 | 4.81 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr20_-_54517709 | 4.52 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr2_+_69415057 | 4.40 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr14_-_6201002 | 4.34 |
ENSRNOT00000049165
|
LOC100366030
|
rCG37858-like |
| chr17_-_21705773 | 4.32 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_-_14367898 | 4.26 |
ENSRNOT00000065402
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr20_-_3440769 | 4.16 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr14_-_6148830 | 4.15 |
ENSRNOT00000075091
ENSRNOT00000092683 |
LOC100366030
|
rCG37858-like |
| chr7_+_141053876 | 4.12 |
ENSRNOT00000084674
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_181103774 | 4.11 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr19_-_53754602 | 3.72 |
ENSRNOT00000035651
|
Fam92b
|
family with sequence similarity 92, member B |
| chr5_-_78324278 | 3.53 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr13_+_78979321 | 3.45 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr1_-_131454689 | 3.37 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_28799266 | 3.36 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr5_-_14356692 | 3.35 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr3_+_151335292 | 3.25 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr2_+_113007549 | 3.20 |
ENSRNOT00000017758
|
Tnfsf10
|
tumor necrosis factor superfamily member 10 |
| chr7_-_70829815 | 3.05 |
ENSRNOT00000011082
|
Shmt2
|
serine hydroxymethyltransferase 2 |
| chr1_+_247238798 | 2.99 |
ENSRNOT00000083511
|
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr1_+_201620642 | 2.97 |
ENSRNOT00000093674
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_+_96449351 | 2.97 |
ENSRNOT00000091272
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr10_-_27179900 | 2.88 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr7_-_70661891 | 2.88 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr12_-_39697465 | 2.87 |
ENSRNOT00000078683
|
Vps29
|
VPS29 retromer complex component |
| chr17_+_43661222 | 2.86 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr4_+_138441332 | 2.81 |
ENSRNOT00000090847
|
Cntn4
|
contactin 4 |
| chr7_-_119071712 | 2.79 |
ENSRNOT00000037611
|
Myh9l1
|
myosin heavy chain 9-like 1 |
| chr10_-_67478848 | 2.77 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr2_+_18392142 | 2.61 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_-_165591110 | 2.53 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr10_-_27179254 | 2.52 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chrX_+_115351114 | 2.48 |
ENSRNOT00000047553
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr18_-_26656879 | 2.35 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr6_+_122603269 | 2.34 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chr3_-_147819571 | 2.25 |
ENSRNOT00000009840
|
Trib3
|
tribbles pseudokinase 3 |
| chr7_+_28414350 | 2.23 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr12_+_22449985 | 2.14 |
ENSRNOT00000075951
|
Slc12a9
|
solute carrier family 12, member 9 |
| chr10_+_53570989 | 2.13 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr10_+_47065850 | 2.05 |
ENSRNOT00000077868
ENSRNOT00000066250 |
Dhrs7b
|
dehydrogenase/reductase 7B |
| chr3_-_122946289 | 1.98 |
ENSRNOT00000055810
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_+_47032113 | 1.98 |
ENSRNOT00000024412
|
Tulp4
|
tubby like protein 4 |
| chr15_-_25505691 | 1.97 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr7_-_9711928 | 1.95 |
ENSRNOT00000011167
|
Neurod4
|
neuronal differentiation 4 |
| chr1_-_169598835 | 1.90 |
ENSRNOT00000072996
|
Olr156
|
olfactory receptor 156 |
| chr4_+_175729726 | 1.86 |
ENSRNOT00000013230
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr5_-_24560063 | 1.81 |
ENSRNOT00000037869
|
Dpy19l4
|
dpy-19-like 4 (C. elegans) |
| chr17_-_53713408 | 1.73 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr11_-_72814850 | 1.71 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chr5_-_28164326 | 1.68 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr14_+_8080275 | 1.66 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr2_+_196013799 | 1.65 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr14_-_28536260 | 1.62 |
ENSRNOT00000059942
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chrX_-_31851715 | 1.61 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr7_+_122818975 | 1.60 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr1_+_61268248 | 1.59 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr6_+_26241672 | 1.53 |
ENSRNOT00000006543
|
Supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr2_+_257568613 | 1.52 |
ENSRNOT00000066125
|
Usp33
|
ubiquitin specific peptidase 33 |
| chr5_+_146337491 | 1.47 |
ENSRNOT00000029395
|
LOC100912566
|
UPF0500 protein C1orf216 homolog |
| chr10_-_109979712 | 1.46 |
ENSRNOT00000086042
|
Dus1l
|
dihydrouridine synthase 1-like |
| chr8_+_117106576 | 1.25 |
ENSRNOT00000071664
|
Rhoa
|
ras homolog family member A |
| chr16_+_74076812 | 1.16 |
ENSRNOT00000025675
|
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr1_+_60248695 | 1.13 |
ENSRNOT00000079558
|
Vom1r9
|
vomeronasal 1 receptor 9 |
| chr14_-_33107776 | 1.06 |
ENSRNOT00000039252
|
Polr2b
|
RNA polymerase II subunit B |
| chr7_-_120770435 | 1.03 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr14_+_39663421 | 1.01 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr6_+_107581608 | 0.98 |
ENSRNOT00000058101
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr1_+_198067066 | 0.97 |
ENSRNOT00000084824
|
Sgf29
|
SAGA complex associated factor 29 |
| chr17_+_57031766 | 0.93 |
ENSRNOT00000092187
ENSRNOT00000068545 |
Crem
|
cAMP responsive element modulator |
| chr10_-_67479077 | 0.90 |
ENSRNOT00000090278
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr2_+_39314714 | 0.85 |
ENSRNOT00000083786
ENSRNOT00000047282 |
Smim15
|
small integral membrane protein 15 |
| chr3_-_74371385 | 0.83 |
ENSRNOT00000041668
|
Olr518
|
olfactory receptor 518 |
| chr16_+_37500017 | 0.83 |
ENSRNOT00000081634
|
AC135696.1
|
high mobility group box 1 (Hmgb1), mRNA |
| chr5_+_145188323 | 0.78 |
ENSRNOT00000038888
|
Tmem35b
|
transmembrane protein 35B |
| chr5_-_77316764 | 0.75 |
ENSRNOT00000071395
ENSRNOT00000076464 |
Mup4
|
major urinary protein 4 |
| chr7_-_123572082 | 0.73 |
ENSRNOT00000068020
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr2_+_44289393 | 0.67 |
ENSRNOT00000018877
|
Il6st
|
interleukin 6 signal transducer |
| chr1_-_167992529 | 0.65 |
ENSRNOT00000067344
|
LOC684170
|
similar to olfactory receptor 566 |
| chr16_+_2379480 | 0.64 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr15_+_61731735 | 0.60 |
ENSRNOT00000015517
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr8_+_18969478 | 0.56 |
ENSRNOT00000061325
|
Olr1130
|
olfactory receptor 1130 |
| chr3_+_167513759 | 0.54 |
ENSRNOT00000072202
|
AABR07054716.1
|
|
| chr3_-_78112770 | 0.52 |
ENSRNOT00000008396
|
Olr694
|
olfactory receptor 694 |
| chr17_+_23661429 | 0.51 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr12_+_38459832 | 0.51 |
ENSRNOT00000090343
|
Vps33a
|
VPS33A CORVET/HOPS core subunit |
| chr6_-_10592454 | 0.48 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr5_+_113592919 | 0.47 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
| chr4_+_92431710 | 0.45 |
ENSRNOT00000049438
|
LOC108350839
|
high mobility group protein B1-like |
| chr11_+_80358211 | 0.44 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr3_+_80670140 | 0.44 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr8_-_17965418 | 0.43 |
ENSRNOT00000072508
|
LOC103693022
|
olfactory receptor 7G2-like |
| chr3_-_74448279 | 0.41 |
ENSRNOT00000043964
|
Olr527
|
olfactory receptor 527 |
| chr1_-_84732242 | 0.38 |
ENSRNOT00000025586
|
LOC102553760
|
zinc finger protein 59-like |
| chr12_+_36638457 | 0.36 |
ENSRNOT00000085692
|
Ubc
|
ubiquitin C |
| chr13_-_74331214 | 0.36 |
ENSRNOT00000006018
|
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr12_-_10973689 | 0.36 |
ENSRNOT00000072406
|
AABR07035357.1
|
|
| chr16_+_8207223 | 0.33 |
ENSRNOT00000026751
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr10_-_60406203 | 0.32 |
ENSRNOT00000041318
|
Olr1505
|
olfactory receptor 1505 |
| chr11_-_1836897 | 0.32 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr1_+_258210344 | 0.31 |
ENSRNOT00000001990
|
LOC100361492
|
cytochrome P450, family 2, subfamily c, polypeptide 55-like |
| chr3_-_73668999 | 0.25 |
ENSRNOT00000080395
|
LOC100909940
|
olfactory receptor 8J3-like |
| chr6_+_136004214 | 0.23 |
ENSRNOT00000013695
|
NEWGENE_619861
|
eukaryotic translation initiation factor 5 |
| chr1_+_86429262 | 0.23 |
ENSRNOT00000045789
|
Vom1r3
|
vomeronasal 1 receptor 3 |
| chr4_-_60548021 | 0.21 |
ENSRNOT00000065546
|
AABR07060165.1
|
|
| chr1_+_43475589 | 0.20 |
ENSRNOT00000092034
ENSRNOT00000024682 |
Oprm1
|
opioid receptor, mu 1 |
| chrX_+_156655960 | 0.19 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr1_+_168204985 | 0.17 |
ENSRNOT00000049036
|
Olr75
|
olfactory receptor 75 |
| chr14_+_106153575 | 0.16 |
ENSRNOT00000010264
|
Vps54
|
VPS54 GARP complex subunit |
| chr8_+_21271190 | 0.16 |
ENSRNOT00000048082
|
LOC100361194
|
olfactory receptor Olr1192-like |
| chr1_-_73732118 | 0.12 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr2_-_96509424 | 0.12 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr15_-_55277713 | 0.09 |
ENSRNOT00000023037
|
Itm2b
|
integral membrane protein 2B |
| chr19_-_38344845 | 0.06 |
ENSRNOT00000072666
|
AABR07043748.1
|
|
| chr1_+_169153297 | 0.04 |
ENSRNOT00000052078
|
Olr137
|
olfactory receptor 137 |
| chr8_+_21305250 | 0.02 |
ENSRNOT00000050206
|
Olr1193
|
olfactory receptor 1193 |
| chr20_+_7330250 | 0.01 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr19_-_27535792 | 0.00 |
ENSRNOT00000024040
|
Olr1666
|
olfactory receptor 1666 |
| chr16_-_61091169 | 0.00 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) positive regulation of lymphangiogenesis(GO:1901492) |
| 2.3 | 6.9 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 2.1 | 12.5 | GO:0046687 | response to chromate(GO:0046687) |
| 1.5 | 5.8 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.4 | 7.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.4 | 4.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.3 | 9.3 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.3 | 6.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.1 | 3.4 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.1 | 7.8 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.0 | 4.8 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 1.0 | 2.9 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.7 | 9.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.7 | 4.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.6 | 1.2 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.6 | 3.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 7.5 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.6 | 6.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.6 | 2.2 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.5 | 1.6 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.5 | 3.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.5 | 1.8 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 5.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 7.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.3 | 3.0 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 4.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 5.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 2.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 2.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 5.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 3.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 2.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 3.2 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.2 | 1.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 4.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.7 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.2 | 0.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 1.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 12.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.2 | 3.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 2.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 | 2.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 14.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 2.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 1.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 4.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 2.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.5 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 1.7 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.2 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 2.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 2.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 5.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 3.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 5.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 1.5 | 7.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.9 | 7.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 4.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.7 | 2.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.6 | 3.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 2.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 2.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 2.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 6.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.2 | 8.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 3.0 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 3.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 7.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.0 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.1 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 11.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 9.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 7.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 10.4 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 8.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 4.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 1.2 | 3.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.2 | 5.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.1 | 4.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.0 | 4.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.0 | 3.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.8 | 6.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.6 | 4.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 5.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 6.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 3.0 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.4 | 2.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.4 | 9.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 5.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 7.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 7.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 9.9 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.7 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 3.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 2.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 12.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 1.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 5.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 7.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 3.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 3.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 7.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 5.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 11.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 16.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.5 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 4.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 10.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 7.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 8.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 5.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 13.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 9.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 11.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.5 | 9.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.4 | 5.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 7.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.4 | 6.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 7.6 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.3 | 4.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 2.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 8.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 2.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 1.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 4.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 7.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 6.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |