Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
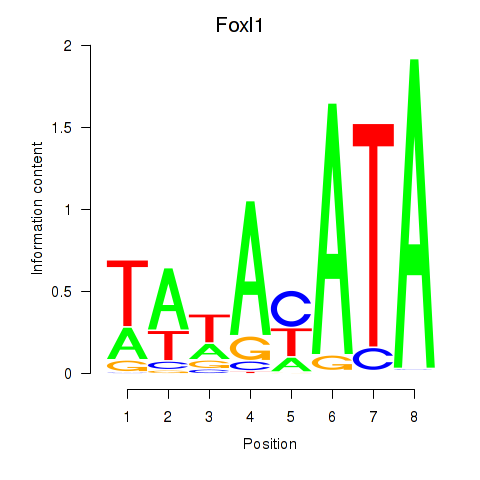
Results for Foxl1
Z-value: 0.28
Transcription factors associated with Foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxl1
|
ENSRNOG00000047125 | forkhead box L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxl1 | rn6_v1_chr19_+_53055745_53055745 | -0.14 | 1.1e-02 | Click! |
Activity profile of Foxl1 motif
Sorted Z-values of Foxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_41212072 | 12.66 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr14_+_26662965 | 11.29 |
ENSRNOT00000002621
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr11_-_32858830 | 9.11 |
ENSRNOT00000002313
|
Runx1
|
runt-related transcription factor 1 |
| chr17_+_43734461 | 4.70 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr17_-_43640387 | 4.36 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr3_+_111135021 | 3.67 |
ENSRNOT00000018970
|
Dll4
|
delta like canonical Notch ligand 4 |
| chr19_-_49448072 | 2.31 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr9_-_71798265 | 1.90 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chr3_+_73344447 | 1.06 |
ENSRNOT00000045693
|
Olr471
|
olfactory receptor 471 |
| chr2_+_209849848 | 1.01 |
ENSRNOT00000086836
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr4_-_51931112 | 0.98 |
ENSRNOT00000080575
|
Pot1
|
protection of telomeres 1 |
| chr3_+_73366155 | 0.36 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
| chr15_-_36321572 | 0.13 |
ENSRNOT00000073934
|
AABR07018016.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.2 | 3.7 | GO:1903588 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.1 | 9.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.7 | 11.3 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.3 | 1.0 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.3 | 1.9 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 1.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 4.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 9.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0032564 | dATP binding(GO:0032564) |
| 0.3 | 9.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.2 | 11.3 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.2 | 3.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 4.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 9.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 3.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |