Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
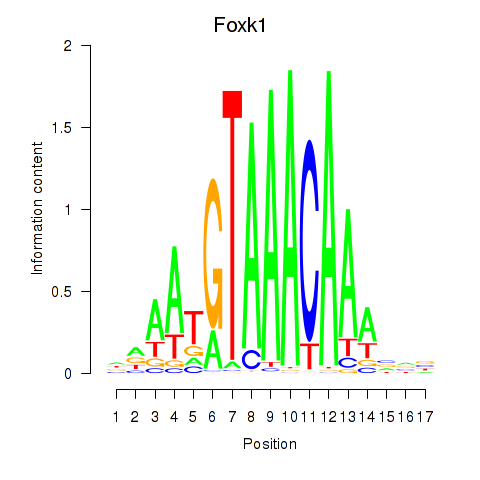
Results for Foxk1_Foxj1
Z-value: 0.66
Transcription factors associated with Foxk1_Foxj1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxk1
|
ENSRNOG00000001104 | forkhead box K1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxk1 | rn6_v1_chr12_-_14244316_14244316 | 0.08 | 1.8e-01 | Click! |
Activity profile of Foxk1_Foxj1 motif
Sorted Z-values of Foxk1_Foxj1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_206282575 | 20.21 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr5_-_136049551 | 15.38 |
ENSRNOT00000068395
ENSRNOT00000079371 |
Kif2c
|
kinesin family member 2C |
| chr2_-_3124543 | 15.08 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr1_+_141488272 | 14.21 |
ENSRNOT00000034042
|
Wdr93
|
WD repeat domain 93 |
| chr5_-_173484986 | 13.65 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr8_-_128711221 | 11.95 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr17_-_15484269 | 11.92 |
ENSRNOT00000020648
|
Omd
|
osteomodulin |
| chr18_-_70924708 | 11.73 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr5_+_103251986 | 11.72 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr3_+_117421604 | 11.39 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr3_-_72219246 | 10.88 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr2_+_208749996 | 10.30 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr10_-_44746549 | 9.26 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr10_+_74724549 | 9.20 |
ENSRNOT00000009077
|
Tex14
|
testis expressed 14, intercellular bridge forming factor |
| chr1_+_256035866 | 9.07 |
ENSRNOT00000079726
|
Kif11
|
kinesin family member 11 |
| chrX_-_65335987 | 8.88 |
ENSRNOT00000047128
|
AABR07038981.1
|
|
| chr18_-_26658892 | 8.81 |
ENSRNOT00000038247
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr17_-_15478343 | 8.71 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr3_+_170996193 | 8.62 |
ENSRNOT00000008959
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr5_-_28131133 | 8.46 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr1_-_156327352 | 8.42 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr1_+_40891597 | 8.37 |
ENSRNOT00000026482
|
Akap12
|
A-kinase anchoring protein 12 |
| chr17_-_8925503 | 8.12 |
ENSRNOT00000015163
|
Catsper3
|
cation channel, sperm associated 3 |
| chr1_+_154606490 | 8.09 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr9_-_65790347 | 8.08 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr2_+_208750356 | 7.57 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr9_+_52023295 | 7.42 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr7_-_140600818 | 7.35 |
ENSRNOT00000082375
|
Lmbr1l
|
limb development membrane protein 1-like |
| chr4_+_52129556 | 7.34 |
ENSRNOT00000009435
|
Hyal6
|
hyaluronoglucosaminidase 6 |
| chr1_-_189238776 | 7.06 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr15_-_41595345 | 6.97 |
ENSRNOT00000019639
|
Sgcg
|
sarcoglycan, gamma |
| chr9_+_10760113 | 6.84 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr10_-_87564327 | 6.68 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr13_-_56462834 | 6.63 |
ENSRNOT00000032122
|
Crb1
|
crumbs 1, cell polarity complex component |
| chr15_-_39573776 | 6.63 |
ENSRNOT00000082517
|
LOC691850
|
hypothetical protein LOC691850 |
| chr4_-_115239723 | 6.51 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr3_+_101791337 | 6.34 |
ENSRNOT00000046937
|
Slc5a12
|
solute carrier family 5 member 12 |
| chr8_+_18430302 | 6.34 |
ENSRNOT00000008341
|
Mbd3l1
|
methyl-CpG binding domain protein 3-like 1 |
| chr20_+_26988774 | 6.24 |
ENSRNOT00000090083
|
Mypn
|
myopalladin |
| chr10_-_93675991 | 6.19 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr4_-_181348038 | 6.16 |
ENSRNOT00000082879
|
LOC690784
|
hypothetical protein LOC690783 |
| chr17_+_32973695 | 6.14 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr2_+_164549455 | 6.08 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr14_-_62595854 | 5.90 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr18_-_45380730 | 5.87 |
ENSRNOT00000037904
|
Pudp
|
pseudouridine 5'-phosphatase |
| chr13_-_108178609 | 5.84 |
ENSRNOT00000004525
|
Cenpf
|
centromere protein F |
| chr5_-_57896475 | 5.78 |
ENSRNOT00000017903
|
RGD1561916
|
similar to testes development-related NYD-SP22 isoform 1 |
| chr10_+_92769031 | 5.74 |
ENSRNOT00000090759
|
AABR07030543.1
|
|
| chr1_-_227486976 | 5.66 |
ENSRNOT00000035812
|
Ms4a14
|
membrane spanning 4-domains A14 |
| chrY_+_914045 | 5.56 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr7_-_107775712 | 5.45 |
ENSRNOT00000010811
ENSRNOT00000093459 |
Ndrg1
|
N-myc downstream regulated 1 |
| chr8_+_79054237 | 5.41 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr2_+_193866951 | 5.39 |
ENSRNOT00000013393
|
S100a11
|
S100 calcium binding protein A11 |
| chr18_+_3887419 | 5.37 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr2_-_165591110 | 5.37 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr2_-_187909394 | 5.36 |
ENSRNOT00000032355
|
Rab25
|
RAB25, member RAS oncogene family |
| chr10_-_97750679 | 5.34 |
ENSRNOT00000078059
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr4_-_155275161 | 5.28 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr9_+_23503236 | 5.14 |
ENSRNOT00000017996
|
Crisp2
|
cysteine-rich secretory protein 2 |
| chr2_-_84531192 | 5.09 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr3_-_119611136 | 4.91 |
ENSRNOT00000016157
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr14_+_23089699 | 4.89 |
ENSRNOT00000036948
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr18_-_15089988 | 4.88 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr17_-_21677477 | 4.86 |
ENSRNOT00000035448
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr10_+_70655818 | 4.85 |
ENSRNOT00000074839
|
LOC689039
|
hypothetical protein LOC689039 |
| chr5_-_78324278 | 4.84 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr5_-_33892462 | 4.80 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr9_+_77320726 | 4.79 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr4_+_56591715 | 4.77 |
ENSRNOT00000041207
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr18_+_63599425 | 4.71 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr17_+_60287203 | 4.62 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chr3_-_148932878 | 4.59 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr1_+_47605262 | 4.55 |
ENSRNOT00000089458
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr5_-_59149625 | 4.50 |
ENSRNOT00000040641
ENSRNOT00000084216 |
Spag8
|
sperm associated antigen 8 |
| chr6_+_27887797 | 4.49 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr2_-_40386669 | 4.49 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr3_+_129462738 | 4.48 |
ENSRNOT00000077755
|
AABR07053830.1
|
|
| chr4_+_30313102 | 4.48 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chrX_+_33443186 | 4.41 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr5_+_162351001 | 4.38 |
ENSRNOT00000065471
|
LOC691162
|
hypothetical protein LOC691162 |
| chr1_+_47605415 | 4.28 |
ENSRNOT00000034842
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr20_+_44060731 | 4.24 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr2_+_143656793 | 4.19 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr3_-_46726946 | 4.16 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr1_+_274049533 | 4.15 |
ENSRNOT00000076779
ENSRNOT00000042822 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr2_-_88414012 | 4.09 |
ENSRNOT00000014762
|
Lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr1_+_91716383 | 4.02 |
ENSRNOT00000016919
|
Slc7a9
|
solute carrier family 7 member 9 |
| chr8_+_70915953 | 3.96 |
ENSRNOT00000046914
|
Rasl12
|
RAS-like, family 12 |
| chr5_-_122828398 | 3.95 |
ENSRNOT00000030112
|
RGD1562532
|
hypothetical gene supported by BC079057 |
| chr1_+_217039755 | 3.94 |
ENSRNOT00000091603
|
LOC102552318
|
actin-like |
| chr1_-_101773508 | 3.92 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr4_+_157348020 | 3.90 |
ENSRNOT00000020803
|
Cdca3
|
cell division cycle associated 3 |
| chr3_-_134462653 | 3.86 |
ENSRNOT00000033562
|
Sel1l2
|
SEL1L2 ERAD E3 ligase adaptor subunit |
| chrX_+_54734385 | 3.85 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr3_-_173944396 | 3.84 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr1_+_142679345 | 3.84 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr4_+_123801174 | 3.84 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr8_+_23398030 | 3.83 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr9_-_65693822 | 3.83 |
ENSRNOT00000038431
|
Als2cr12
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 12 |
| chr7_+_58814805 | 3.81 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr2_+_197682000 | 3.80 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chrX_+_35869538 | 3.79 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chrY_-_1398030 | 3.77 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chrX_-_24120987 | 3.77 |
ENSRNOT00000030377
|
LOC685762
|
hypothetical protein LOC685762 |
| chr15_+_2526368 | 3.75 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr3_-_119247449 | 3.73 |
ENSRNOT00000015238
|
Usp50
|
ubiquitin specific peptidase 50 |
| chr20_+_29655226 | 3.70 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr4_+_51614676 | 3.67 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_-_130475320 | 3.65 |
ENSRNOT00000075439
|
Ccdc13
|
coiled-coil domain containing 13 |
| chrX_-_16306078 | 3.62 |
ENSRNOT00000003939
|
Akap4
|
A-kinase anchoring protein 4 |
| chr10_+_49472460 | 3.62 |
ENSRNOT00000038276
|
Tekt3
|
tektin 3 |
| chr7_+_117326279 | 3.61 |
ENSRNOT00000050522
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr17_-_14637832 | 3.60 |
ENSRNOT00000074323
|
LOC103690116
|
osteomodulin |
| chr15_+_30853161 | 3.57 |
ENSRNOT00000086722
|
AABR07017768.4
|
|
| chr6_-_99273033 | 3.57 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr1_-_252550394 | 3.55 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_+_53013413 | 3.54 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr17_-_30661690 | 3.53 |
ENSRNOT00000048146
|
RGD1307537
|
similar to RIKEN cDNA 4933417A18 |
| chr5_-_50075454 | 3.52 |
ENSRNOT00000011085
|
Orc3
|
origin recognition complex, subunit 3 |
| chr8_-_78397123 | 3.52 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr4_-_129619142 | 3.50 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chrX_-_77559348 | 3.47 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr2_+_116416507 | 3.47 |
ENSRNOT00000030700
|
Actrt3
|
actin-related protein T3 |
| chr8_-_87213627 | 3.47 |
ENSRNOT00000066084
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr5_+_145188323 | 3.46 |
ENSRNOT00000038888
|
Tmem35b
|
transmembrane protein 35B |
| chr6_-_23291568 | 3.43 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_-_56269242 | 3.43 |
ENSRNOT00000021901
|
Lyzl1
|
lysozyme-like 1 |
| chr20_-_5037022 | 3.42 |
ENSRNOT00000091022
ENSRNOT00000060762 |
Msh5
|
mutS homolog 5 |
| chr7_-_12346475 | 3.41 |
ENSRNOT00000060708
|
Mum1
|
melanoma associated antigen (mutated) 1 |
| chr16_-_47207487 | 3.39 |
ENSRNOT00000059459
|
Dctd
|
dCMP deaminase |
| chr8_-_84320714 | 3.38 |
ENSRNOT00000079356
ENSRNOT00000088487 |
Tinag
|
tubulointerstitial nephritis antigen |
| chr8_-_61519507 | 3.36 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chr17_-_15467320 | 3.34 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr1_-_7480825 | 3.30 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr1_+_7252349 | 3.30 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr3_+_11653529 | 3.27 |
ENSRNOT00000091048
|
Ak1
|
adenylate kinase 1 |
| chr1_+_201913148 | 3.25 |
ENSRNOT00000036128
|
Fam24a
|
family with sequence similarity 24, member A |
| chr4_+_147756553 | 3.24 |
ENSRNOT00000086549
ENSRNOT00000014733 |
Ift122
|
intraflagellar transport 122 |
| chr10_+_59894340 | 3.22 |
ENSRNOT00000080446
|
Spata22
|
spermatogenesis associated 22 |
| chr10_+_13214130 | 3.22 |
ENSRNOT00000073065
|
Prss21
|
protease, serine, 21 |
| chr5_-_56567320 | 3.17 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr14_-_82171480 | 3.14 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr19_-_41349681 | 3.13 |
ENSRNOT00000080694
ENSRNOT00000059147 |
Hydin
|
Hydin, axonemal central pair apparatus protein |
| chr13_+_56546021 | 3.13 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr16_+_72216326 | 3.12 |
ENSRNOT00000051363
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr2_+_236233239 | 3.12 |
ENSRNOT00000013694
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr17_+_47302272 | 3.12 |
ENSRNOT00000090021
|
Nme8
|
NME/NM23 family member 8 |
| chr10_-_34990943 | 3.11 |
ENSRNOT00000075555
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr7_+_120580743 | 3.08 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr10_-_41491554 | 3.07 |
ENSRNOT00000035759
|
LOC102552619
|
uncharacterized LOC102552619 |
| chr4_-_144620731 | 3.07 |
ENSRNOT00000066468
|
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr3_+_155269347 | 3.04 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr11_+_45339366 | 3.04 |
ENSRNOT00000002241
|
Tmem30c
|
transmembrane protein 30C |
| chr5_-_166282831 | 3.03 |
ENSRNOT00000021348
|
Rbp7
|
retinol binding protein 7 |
| chr16_-_61091169 | 3.02 |
ENSRNOT00000016328
|
Dusp4
|
dual specificity phosphatase 4 |
| chr1_+_162890475 | 3.00 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr6_+_64649194 | 3.00 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr2_-_203350736 | 2.98 |
ENSRNOT00000090115
ENSRNOT00000084516 |
Ttf2
|
transcription termination factor 2 |
| chr15_-_70349983 | 2.98 |
ENSRNOT00000012167
|
Diaph3
|
diaphanous-related formin 3 |
| chr3_+_101899068 | 2.97 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr1_-_20417637 | 2.94 |
ENSRNOT00000036049
|
RGD1559962
|
similar to High mobility group protein 2 (HMG-2) |
| chr13_+_97838361 | 2.93 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chrX_+_106791333 | 2.92 |
ENSRNOT00000050302
|
Tceal9
|
transcription elongation factor A like 9 |
| chr2_+_240396152 | 2.92 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr2_+_260039651 | 2.91 |
ENSRNOT00000073873
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr5_-_153737161 | 2.91 |
ENSRNOT00000073506
|
Ncmap
|
noncompact myelin associated protein |
| chr6_-_99843245 | 2.88 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr20_+_18780605 | 2.87 |
ENSRNOT00000000754
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr12_+_30603242 | 2.87 |
ENSRNOT00000068460
|
Sept14
|
septin 14 |
| chr1_-_48360131 | 2.87 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr3_-_11820549 | 2.86 |
ENSRNOT00000020660
|
Cfap157
|
cilia and flagella associated protein 157 |
| chr1_-_60407295 | 2.84 |
ENSRNOT00000078350
|
Vom1r12
|
vomeronasal 1 receptor 12 |
| chr1_+_225068009 | 2.84 |
ENSRNOT00000026651
|
Ubxn1
|
UBX domain protein 1 |
| chr10_-_109267500 | 2.83 |
ENSRNOT00000005977
|
Cep131
|
centrosomal protein 131 |
| chr8_-_127871192 | 2.82 |
ENSRNOT00000055928
|
Slc22a14
|
solute carrier family 22, member 14 |
| chr14_-_44078897 | 2.78 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr5_-_17399801 | 2.77 |
ENSRNOT00000011900
|
RGD1563405
|
similar to protein tyrosine phosphatase 4a2 |
| chr10_-_4910305 | 2.75 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chrX_+_119390013 | 2.75 |
ENSRNOT00000074269
|
Agtr2
|
angiotensin II receptor, type 2 |
| chr2_+_264704769 | 2.75 |
ENSRNOT00000012667
|
Depdc1
|
DEP domain containing 1 |
| chr4_-_61502778 | 2.74 |
ENSRNOT00000051232
|
AABR07060190.1
|
|
| chr1_-_69835185 | 2.70 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr12_-_47653638 | 2.69 |
ENSRNOT00000001577
|
Tchp
|
trichoplein, keratin filament binding |
| chr3_-_8979889 | 2.68 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr16_-_47208110 | 2.67 |
ENSRNOT00000017670
|
Dctd
|
dCMP deaminase |
| chr11_-_68349238 | 2.67 |
ENSRNOT00000030074
|
Sema5b
|
semaphorin 5B |
| chrX_+_23414354 | 2.66 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr15_+_41937880 | 2.64 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr8_+_103938520 | 2.63 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr5_-_146069670 | 2.61 |
ENSRNOT00000072793
|
LOC682102
|
hypothetical protein LOC682102 |
| chr2_+_193724248 | 2.60 |
ENSRNOT00000025249
|
Rptn
|
repetin |
| chr17_+_81922329 | 2.60 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr9_+_76913639 | 2.58 |
ENSRNOT00000089203
|
AABR07068007.2
|
|
| chr2_+_154921999 | 2.58 |
ENSRNOT00000057620
|
LOC691044
|
similar to GTPase activating protein testicular GAP1 |
| chrX_-_45000363 | 2.57 |
ENSRNOT00000035538
|
Cldn34e
|
claudin 34E |
| chr17_+_14469488 | 2.57 |
ENSRNOT00000060670
|
AABR07027111.1
|
|
| chrX_+_120624518 | 2.55 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr3_+_112531703 | 2.55 |
ENSRNOT00000041727
|
LOC100911204
|
protein CASC5-like |
| chr9_-_44419998 | 2.55 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr10_-_37084637 | 2.54 |
ENSRNOT00000075525
|
LOC103689927
|
protein RMD5 homolog B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxk1_Foxj1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 3.9 | 11.7 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 3.8 | 11.4 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.6 | 17.9 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 2.6 | 15.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.2 | 11.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 2.0 | 10.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.9 | 7.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.6 | 4.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.5 | 9.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 1.4 | 4.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.2 | 3.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.1 | 9.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 1.1 | 12.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 1.1 | 3.3 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 1.0 | 3.1 | GO:0003285 | septum secundum development(GO:0003285) |
| 1.0 | 3.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.0 | 6.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 1.0 | 7.1 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 1.0 | 4.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 1.0 | 2.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.9 | 1.9 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.9 | 2.7 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of metanephros size(GO:0035566) |
| 0.9 | 0.9 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.9 | 4.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.9 | 4.5 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 2.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.8 | 8.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.8 | 4.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.8 | 2.4 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.8 | 3.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.8 | 3.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.8 | 1.5 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.8 | 5.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.7 | 4.5 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.7 | 2.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.7 | 2.9 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.7 | 2.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.7 | 2.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.7 | 3.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.7 | 2.8 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.7 | 3.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.7 | 2.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.7 | 2.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 2.0 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.7 | 2.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.7 | 8.5 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.6 | 1.9 | GO:0071038 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.6 | 3.8 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.6 | 1.9 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.6 | 1.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.6 | 2.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.6 | 1.8 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.6 | 5.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.6 | 3.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.6 | 5.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.6 | 2.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.5 | 1.6 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.5 | 5.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.5 | 7.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.5 | 2.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.5 | 1.6 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.5 | 3.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.5 | 3.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.5 | 6.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 1.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.5 | 2.1 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.5 | 13.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.5 | 7.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 3.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.5 | 1.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.5 | 2.9 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.5 | 5.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.5 | 1.4 | GO:0016487 | response to jasmonic acid(GO:0009753) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of isoprenoid metabolic process(GO:0019747) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.5 | 1.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 3.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 1.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.5 | 3.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.5 | 1.8 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.4 | 1.8 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 2.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.4 | 3.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 2.2 | GO:0006566 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.4 | 2.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.4 | 0.9 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 2.1 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.4 | 2.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 0.8 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) |
| 0.4 | 8.8 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.4 | 1.2 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.4 | 2.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 2.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 | 1.2 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.4 | 1.1 | GO:0090024 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) cellular response to heparin(GO:0071504) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.4 | 1.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.4 | 5.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.4 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.4 | 1.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 2.9 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 1.1 | GO:0072134 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) gonad morphogenesis(GO:0035262) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.3 | 6.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 1.3 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 3.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 2.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.3 | 1.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 1.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.5 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.3 | 21.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.3 | 1.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 0.9 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 6.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.3 | 3.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.3 | 5.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.9 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 0.5 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.3 | 0.8 | GO:1905065 | regulation of cell growth by extracellular stimulus(GO:0001560) positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.3 | 2.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 2.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 0.8 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.3 | 3.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.3 | 10.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.3 | 0.8 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) cellular response to astaxanthin(GO:1905218) |
| 0.3 | 2.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 1.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.7 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 4.5 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 9.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 6.5 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.2 | 1.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 3.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 3.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 1.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 2.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 2.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 5.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 0.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 2.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 3.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 1.8 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 1.8 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 7.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 1.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 4.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 1.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.7 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.2 | 1.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 1.8 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 2.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 1.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 2.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 2.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 2.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 11.3 | GO:0048644 | muscle organ morphogenesis(GO:0048644) |
| 0.2 | 14.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.2 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 3.8 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 2.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 2.9 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 0.9 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.2 | 7.4 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.2 | 0.5 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.1 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 1.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 3.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.4 | GO:0045401 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 2.1 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.8 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) sequestering of BMP in extracellular matrix(GO:0035582) determination of dorsal identity(GO:0048263) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 | 0.4 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.8 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 2.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 2.0 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.7 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 2.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.4 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.8 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 4.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) baculum development(GO:1990375) |
| 0.1 | 1.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 1.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.7 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 3.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 3.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 3.3 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 10.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 3.2 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 0.3 | GO:0051542 | negative regulation of iron ion transmembrane transport(GO:0034760) elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 1.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 1.3 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 2.3 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 5.9 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 7.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.0 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 1.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 3.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 2.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 22.7 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 1.6 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 2.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 1.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 4.0 | GO:0018393 | internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 4.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 2.5 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 2.0 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.7 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 10.0 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.4 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.2 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.4 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.8 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.5 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.8 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.5 | 10.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.5 | 3.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.4 | 5.7 | GO:0034657 | GID complex(GO:0034657) |
| 1.3 | 6.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.2 | 3.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.2 | 7.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.0 | 3.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.9 | 13.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.9 | 5.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.8 | 8.6 | GO:0002177 | manchette(GO:0002177) |
| 0.7 | 12.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 5.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.7 | 8.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.6 | 4.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 7.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 4.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 2.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.6 | 4.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 3.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 6.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 14.6 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.4 | 10.9 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 7.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.4 | 4.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 1.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 0.7 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.4 | 1.1 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.4 | 1.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.4 | 1.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 1.5 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.4 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 3.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 3.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 1.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 3.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 2.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 2.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 8.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 2.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 6.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.3 | 0.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 26.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 1.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.3 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 13.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 1.8 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.3 | 3.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 3.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 0.8 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.2 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 9.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 20.5 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 21.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 3.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 10.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 20.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 2.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 1.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 4.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 2.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 1.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 3.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 4.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 13.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 3.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 10.6 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 5.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 4.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 4.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 2.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 5.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 17.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 4.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 13.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.7 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0000803 | sex chromosome(GO:0000803) XY body(GO:0001741) |
| 0.0 | 1.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 10.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 5.8 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 3.8 | 11.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 3.0 | 17.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 2.2 | 8.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.0 | 12.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.8 | 5.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 1.6 | 8.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 1.5 | 6.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 3.9 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 1.2 | 3.6 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.1 | 2.2 | GO:0070905 | serine binding(GO:0070905) |
| 1.0 | 3.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.0 | 3.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.0 | 4.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 1.0 | 6.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.0 | 3.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.0 | 2.9 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.9 | 2.8 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.9 | 7.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.8 | 2.4 | GO:0003922 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.8 | 14.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 3.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.7 | 2.9 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.7 | 7.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.7 | 2.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.7 | 2.1 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.7 | 15.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.7 | 5.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.6 | 9.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 1.9 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.6 | 1.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.6 | 2.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.6 | 2.3 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 2.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.6 | 2.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.6 | 8.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 2.7 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.5 | 7.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 1.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.5 | 1.5 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.5 | 3.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.5 | 1.4 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 1.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 1.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 8.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 2.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.4 | 1.6 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 1.2 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.4 | 2.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.4 | 6.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 2.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 1.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 3.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 7.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 7.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 2.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 4.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 5.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 6.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 2.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 1.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.3 | 4.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 2.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 1.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 3.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 3.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 1.9 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.3 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 3.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 5.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 5.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 3.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 6.1 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 5.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 3.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 6.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 2.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 0.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 5.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 20.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.2 | 1.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.2 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 2.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 2.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 3.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.8 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 2.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 12.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 3.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.6 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.2 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.0 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 3.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 10.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 8.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.1 | 3.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 1.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 4.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 2.8 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 2.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 3.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 1.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 3.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 3.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 2.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 3.7 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.7 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 4.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 4.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 1.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 2.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.2 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 10.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 4.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 3.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 5.2 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 2.0 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 19.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 1.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.5 | 20.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 20.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 1.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 5.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 10.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 3.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 8.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 10.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 24.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.8 | 24.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 5.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 5.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 10.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 2.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 4.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 7.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 2.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 14.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 10.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 3.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 20.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 21.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 3.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 20.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.7 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.4 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 3.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.3 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 5.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |