Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Foxg1
Z-value: 0.59
Transcription factors associated with Foxg1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxg1
|
ENSRNOG00000047891 | forkhead box G1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxg1 | rn6_v1_chr6_+_69971227_69971227 | 0.80 | 1.1e-73 | Click! |
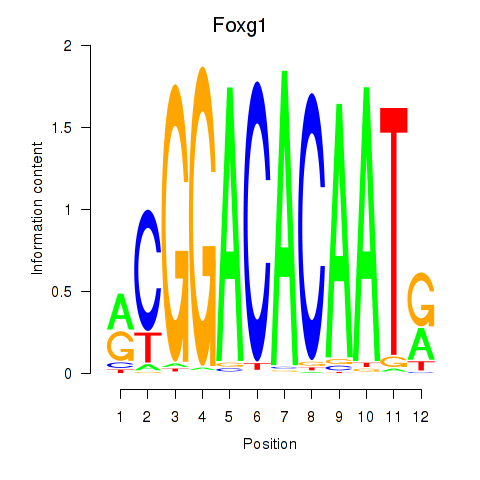
Activity profile of Foxg1 motif
Sorted Z-values of Foxg1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_136853957 | 28.35 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr7_-_136853154 | 26.09 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr2_-_231521052 | 23.46 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr13_-_95348913 | 20.67 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr5_+_172364421 | 19.03 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr9_+_6970507 | 15.48 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr3_-_52664209 | 14.74 |
ENSRNOT00000065126
ENSRNOT00000079020 |
Scn9a
|
sodium voltage-gated channel alpha subunit 9 |
| chr2_+_115739813 | 14.54 |
ENSRNOT00000085452
|
Slc7a14
|
solute carrier family 7, member 14 |
| chrX_+_6273733 | 14.35 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr4_-_7228675 | 12.92 |
ENSRNOT00000064386
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr7_-_126465723 | 12.81 |
ENSRNOT00000021196
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr1_-_219532609 | 11.38 |
ENSRNOT00000025641
|
Ankrd13d
|
ankyrin repeat domain 13D |
| chr5_+_64326733 | 11.37 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr14_+_17534412 | 10.77 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr5_-_20189721 | 10.25 |
ENSRNOT00000014661
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr14_-_112946204 | 9.94 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr10_-_96015499 | 7.88 |
ENSRNOT00000004383
|
Cacng4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr13_-_95250235 | 7.72 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr1_+_81373340 | 7.41 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr15_-_108120279 | 7.21 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr18_+_64084795 | 5.19 |
ENSRNOT00000022410
|
Rnmt
|
RNA (guanine-7-) methyltransferase |
| chr8_-_73194837 | 5.00 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr10_-_84847857 | 4.51 |
ENSRNOT00000071495
|
LOC102552549
|
migration and invasion enhancer 1-like |
| chr8_+_59164572 | 3.87 |
ENSRNOT00000015102
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr10_-_86393141 | 2.90 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr8_+_11931767 | 2.81 |
ENSRNOT00000087963
|
Maml2
|
mastermind-like transcriptional coactivator 2 |
| chr9_-_80167033 | 2.57 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr6_-_111477090 | 2.55 |
ENSRNOT00000016389
|
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr1_+_40389638 | 2.49 |
ENSRNOT00000021471
|
Plekhg1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr18_+_44737154 | 2.41 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr5_+_135574172 | 2.20 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr3_-_22500765 | 2.00 |
ENSRNOT00000025443
|
Dennd1a
|
DENN domain containing 1A |
| chr1_-_164659992 | 1.88 |
ENSRNOT00000024281
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr10_-_18506337 | 1.76 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr5_+_160356245 | 1.66 |
ENSRNOT00000085378
|
Casp9
|
caspase 9 |
| chr8_+_114866768 | 1.61 |
ENSRNOT00000076731
|
Wdr82
|
WD repeat domain 82 |
| chr17_+_77601877 | 1.60 |
ENSRNOT00000091554
ENSRNOT00000024872 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr3_-_148312791 | 1.43 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr1_-_99135977 | 1.34 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr8_-_48727154 | 1.18 |
ENSRNOT00000088471
|
Trappc4
|
trafficking protein particle complex 4 |
| chr6_-_29975730 | 1.14 |
ENSRNOT00000075574
|
Wdcp
|
WD repeat and coiled coil containing |
| chr1_-_92119951 | 0.99 |
ENSRNOT00000018153
ENSRNOT00000092121 |
Zfp507
|
zinc finger protein 507 |
| chr1_-_101236065 | 0.81 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr6_+_111476768 | 0.76 |
ENSRNOT00000016479
|
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr14_-_106864892 | 0.71 |
ENSRNOT00000090664
|
Otx1
|
orthodenticle homeobox 1 |
| chr8_+_114867062 | 0.58 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr4_-_106983279 | 0.55 |
ENSRNOT00000007700
|
AABR07061152.1
|
|
| chr15_+_26887767 | 0.49 |
ENSRNOT00000016674
|
Olr1627
|
olfactory receptor 1627 |
| chr13_+_92264231 | 0.31 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr4_+_68653369 | 0.08 |
ENSRNOT00000046917
|
Tas2r137
|
taste receptor, type 2, member 137 |
| chr20_+_11588200 | 0.07 |
ENSRNOT00000041259
|
AABR07072821.1
|
|
| chr1_+_171188133 | 0.07 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxg1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 19.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 5.9 | 23.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 4.5 | 54.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 4.3 | 12.8 | GO:0072054 | chemoattraction of axon(GO:0061642) renal outer medulla development(GO:0072054) |
| 3.7 | 14.7 | GO:0043179 | rhythmic excitation(GO:0043179) |
| 2.4 | 14.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.3 | 5.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 1.2 | 28.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.9 | 2.6 | GO:0042245 | RNA repair(GO:0042245) |
| 0.7 | 15.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.6 | 3.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.6 | 2.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.5 | 1.6 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.5 | 1.4 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.5 | 1.9 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.5 | 2.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.5 | 5.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 7.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 2.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 12.9 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.2 | 0.8 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.2 | 14.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 0.8 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.7 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 2.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 10.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 2.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 2.4 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 2.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 10.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.7 | 14.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 23.5 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 2.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 1.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 5.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 5.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 7.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 54.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 15.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 2.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 2.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.3 | GO:0032437 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.1 | 1.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 2.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 25.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 3.9 | 15.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.3 | 82.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.0 | 3.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 5.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.6 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.6 | 14.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.5 | 1.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.5 | 23.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 10.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 14.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 2.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 12.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 7.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 14.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 9.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 10.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 5.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 28.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 12.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 57.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 7.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 19.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 3.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 28.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 21.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 7.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 3.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 5.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 12.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |