Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
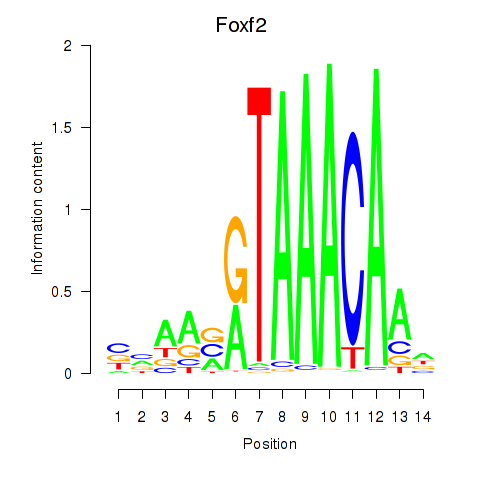
Results for Foxf2
Z-value: 0.86
Transcription factors associated with Foxf2
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Foxf2 motif
Sorted Z-values of Foxf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_89597138 | 56.37 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr18_-_24929091 | 44.51 |
ENSRNOT00000019596
|
Proc
|
protein C |
| chr10_+_56662242 | 43.62 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr8_-_115981910 | 42.42 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr11_-_81660395 | 32.96 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr3_-_101474890 | 32.70 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr2_-_182035032 | 32.63 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr18_+_59748444 | 32.60 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr7_-_119689938 | 32.13 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr7_+_93975451 | 29.99 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr5_+_124300477 | 29.93 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr13_-_80862963 | 26.22 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr5_-_19368431 | 25.80 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr1_+_264741911 | 25.51 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr2_-_100249811 | 22.37 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr3_-_168018410 | 21.58 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr4_-_176528110 | 20.66 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr10_+_103395511 | 20.11 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr10_-_71849293 | 19.90 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr1_-_90978907 | 18.12 |
ENSRNOT00000072819
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr1_+_88750462 | 16.85 |
ENSRNOT00000028247
|
LOC108348122
|
CAP-Gly domain-containing linker protein 3 |
| chr12_-_10391270 | 16.64 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr3_+_142383278 | 16.01 |
ENSRNOT00000017742
|
Foxa2
|
forkhead box A2 |
| chr15_-_28313841 | 15.68 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr5_+_120340646 | 15.08 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr4_+_153921115 | 14.81 |
ENSRNOT00000018821
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr6_-_108167185 | 14.35 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr8_+_94686938 | 14.11 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr3_+_5519990 | 13.89 |
ENSRNOT00000070873
ENSRNOT00000007640 |
Adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr1_+_154606490 | 13.64 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_+_21767606 | 13.41 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr4_+_152630469 | 13.22 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr1_-_156327352 | 12.92 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr5_-_173484986 | 12.44 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr1_+_72545596 | 12.19 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr8_+_52127632 | 12.19 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr1_+_162890475 | 12.00 |
ENSRNOT00000034886
|
Gdpd4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr5_-_114940364 | 11.90 |
ENSRNOT00000038196
|
AABR07049299.1
|
|
| chr9_+_118849302 | 11.88 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr1_-_101773508 | 11.35 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr14_-_21127868 | 11.17 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr8_+_52127399 | 11.06 |
ENSRNOT00000052392
|
Cadm1
|
cell adhesion molecule 1 |
| chr19_-_11669578 | 10.88 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr4_+_57855416 | 10.60 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr2_-_3124543 | 10.28 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr8_-_78233430 | 9.99 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr13_+_26172243 | 9.76 |
ENSRNOT00000003840
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr3_+_60024013 | 9.54 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr9_+_94702129 | 8.86 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr2_+_200603426 | 7.81 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr18_+_30004565 | 7.69 |
ENSRNOT00000027393
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_173944396 | 7.46 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr3_-_160038078 | 7.34 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr14_-_8432195 | 7.29 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr12_-_13998172 | 6.89 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr15_+_39779648 | 5.86 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr8_+_99568958 | 5.84 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr10_+_72909550 | 5.61 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr9_-_65790347 | 5.31 |
ENSRNOT00000028506
|
AABR07067812.1
|
|
| chr6_+_122603269 | 5.29 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chrX_+_65040775 | 5.22 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr13_-_98480419 | 4.88 |
ENSRNOT00000086306
|
Coq8a
|
coenzyme Q8A |
| chr7_-_57816621 | 4.77 |
ENSRNOT00000090748
|
AABR07057150.1
|
|
| chr1_+_214562897 | 4.60 |
ENSRNOT00000085125
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr18_+_13386133 | 4.55 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chrX_+_65040934 | 4.15 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr7_+_28654733 | 4.13 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr7_-_107775712 | 3.77 |
ENSRNOT00000010811
ENSRNOT00000093459 |
Ndrg1
|
N-myc downstream regulated 1 |
| chr13_-_97838228 | 3.65 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr8_+_5522739 | 3.59 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr14_-_6793558 | 3.38 |
ENSRNOT00000002927
|
Mepe
|
matrix extracellular phosphoglycoprotein |
| chr14_-_21810314 | 3.35 |
ENSRNOT00000002671
|
Csn3
|
casein kappa |
| chr10_+_105796680 | 3.18 |
ENSRNOT00000000264
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr3_+_126335863 | 3.02 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr11_+_69739384 | 2.95 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_108167716 | 2.75 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr9_+_111249351 | 2.52 |
ENSRNOT00000076729
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr5_+_50075527 | 2.38 |
ENSRNOT00000011688
|
Rars2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr1_+_190671696 | 2.28 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr18_+_68408890 | 2.03 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr10_+_11393103 | 1.91 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr6_+_91595823 | 1.90 |
ENSRNOT00000006223
|
Klhdc2
|
kelch domain containing 2 |
| chr1_-_214455039 | 1.40 |
ENSRNOT00000071028
|
Polr2l
|
RNA polymerase II subunit L |
| chrX_+_83926513 | 1.24 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr20_-_11721838 | 1.16 |
ENSRNOT00000001636
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr17_+_86408151 | 0.99 |
ENSRNOT00000022734
|
Otud1
|
OTU deubiquitinase 1 |
| chr14_-_3351553 | 0.96 |
ENSRNOT00000061556
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr1_-_102970950 | 0.72 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr2_+_193724248 | 0.51 |
ENSRNOT00000025249
|
Rptn
|
repetin |
| chr3_+_137154086 | 0.41 |
ENSRNOT00000034252
|
Otor
|
otoraplin |
| chr6_-_109095557 | 0.24 |
ENSRNOT00000033493
|
Mlh3
|
mutL homolog 3 |
| chr4_+_71512695 | 0.11 |
ENSRNOT00000037220
|
Tas2r139
|
taste receptor, type 2, member 139 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.8 | 56.4 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 10.9 | 32.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 7.4 | 44.5 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 6.5 | 32.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 6.0 | 35.9 | GO:0001705 | ectoderm formation(GO:0001705) |
| 5.4 | 32.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 4.8 | 14.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 4.7 | 23.3 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 4.4 | 35.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 4.1 | 12.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 3.9 | 15.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 3.8 | 15.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 3.7 | 14.8 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 3.3 | 9.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 2.9 | 32.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 2.7 | 29.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.2 | 8.9 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 2.0 | 25.8 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 1.9 | 20.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.8 | 7.3 | GO:0009597 | detection of virus(GO:0009597) |
| 1.6 | 11.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.5 | 11.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.3 | 14.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 1.2 | 7.5 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 1.0 | 4.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.0 | 3.0 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) embryonic heart tube anterior/posterior pattern specification(GO:0035054) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.9 | 16.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.8 | 29.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.7 | 7.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.7 | 26.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.7 | 3.6 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.7 | 5.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.6 | 2.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 2.9 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.6 | 13.9 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.5 | 11.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.5 | 12.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 3.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.4 | 5.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 5.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.3 | 4.9 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 3.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.7 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 6.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 10.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 3.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 13.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 43.6 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.1 | 2.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 1.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 21.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 28.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 3.3 | GO:0007595 | lactation(GO:0007595) |
| 0.1 | 7.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 56.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 4.3 | 29.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 4.1 | 32.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.5 | 23.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.2 | 35.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 32.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.6 | 11.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 6.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.4 | 30.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.4 | 2.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 20.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 12.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 7.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 4.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 3.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 10.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 17.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 10.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 48.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 3.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 16.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 1.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 23.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 3.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 52.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 11.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 39.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 5.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 7.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.8 | 56.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 14.8 | 44.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 10.9 | 32.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 5.0 | 35.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 4.1 | 12.4 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 3.8 | 11.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 2.7 | 33.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 2.6 | 20.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.5 | 14.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 2.2 | 8.9 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 2.2 | 26.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.0 | 12.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.8 | 10.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.5 | 30.0 | GO:0005537 | mannose binding(GO:0005537) |
| 1.2 | 3.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.0 | 25.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.9 | 11.9 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.7 | 9.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.7 | 32.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.6 | 2.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.6 | 7.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 2.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 14.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.5 | 6.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 3.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 10.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 16.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 3.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.3 | 30.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 21.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 5.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 15.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 18.3 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.2 | 45.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 35.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 3.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 10.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 4.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 4.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 26.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 77.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 10.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 55.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 16.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 2.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 77.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 3.3 | 56.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.4 | 9.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 2.1 | 20.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.9 | 32.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 1.7 | 25.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.4 | 38.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.9 | 29.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.9 | 11.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.8 | 23.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 14.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 14.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 15.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.5 | 26.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.4 | 10.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.4 | 5.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.4 | 4.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 8.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 21.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 31.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 7.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 10.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 4.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |