Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
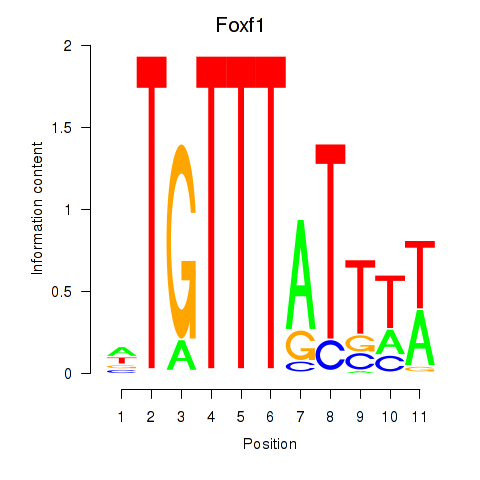
Results for Foxf1
Z-value: 1.01
Transcription factors associated with Foxf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf1
|
ENSRNOG00000049906 | forkhead box F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | rn6_v1_chr19_+_53012332_53012332 | 0.16 | 5.3e-03 | Click! |
Activity profile of Foxf1 motif
Sorted Z-values of Foxf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_253000760 | 31.73 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr5_-_7874909 | 28.43 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr3_+_41019898 | 26.12 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr13_-_47377703 | 23.16 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chr8_-_26345754 | 21.13 |
ENSRNOT00000091773
|
AC094212.1
|
triosephosphate isomerase |
| chr13_+_78805347 | 19.98 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr11_-_81660395 | 19.27 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr15_-_28313841 | 18.40 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr8_-_128711221 | 18.09 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr2_+_200572502 | 17.47 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr1_+_48273611 | 17.33 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr10_-_98294522 | 17.24 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr16_-_81980027 | 16.40 |
ENSRNOT00000043170
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr8_+_57886168 | 16.28 |
ENSRNOT00000039336
|
Exph5
|
exophilin 5 |
| chr4_+_139670092 | 15.88 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr2_+_200603426 | 15.25 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr18_+_68408890 | 14.46 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr9_-_20195566 | 14.39 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr17_+_32973695 | 14.21 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr3_-_162059524 | 14.02 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr8_-_78233430 | 14.00 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrX_-_142248369 | 13.99 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_+_51614676 | 13.98 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr4_+_163293724 | 13.97 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chrX_-_116792864 | 13.97 |
ENSRNOT00000090918
|
Amot
|
angiomotin |
| chr1_-_262047332 | 13.60 |
ENSRNOT00000067274
|
Hpse2
|
heparanase 2 (inactive) |
| chr20_+_26999795 | 13.51 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr8_-_49109981 | 13.49 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr5_+_58636083 | 12.85 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr4_+_9981958 | 12.71 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr2_-_258997138 | 12.30 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr1_-_148119857 | 12.14 |
ENSRNOT00000040325
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr6_-_127534247 | 11.99 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr13_-_56763981 | 11.81 |
ENSRNOT00000087916
|
LOC100361907
|
complement factor H-related protein B |
| chr8_-_115981910 | 11.73 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr2_-_250923744 | 11.63 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr13_+_89597138 | 11.58 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr7_+_93975451 | 11.37 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr19_-_11669578 | 11.11 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr15_-_110612681 | 10.95 |
ENSRNOT00000041551
|
Fgf14
|
fibroblast growth factor 14 |
| chr3_+_149624712 | 10.77 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr1_-_220165678 | 10.73 |
ENSRNOT00000026915
|
Bbs1
|
Bardet-Biedl syndrome 1 |
| chr4_+_181103774 | 10.52 |
ENSRNOT00000084207
ENSRNOT00000055473 ENSRNOT00000077619 |
Arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr3_-_60611924 | 10.42 |
ENSRNOT00000068745
|
Chn1
|
chimerin 1 |
| chr9_+_118849302 | 10.27 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr3_-_51612397 | 10.05 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr4_-_176294997 | 9.97 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr7_+_141370491 | 9.94 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr3_+_140024043 | 9.94 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr1_-_154111725 | 9.92 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr1_-_167911961 | 9.91 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr5_-_173484986 | 9.84 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr7_-_118396728 | 9.81 |
ENSRNOT00000066431
|
Rbfox2
|
RNA binding protein, fox-1 homolog 2 |
| chr15_-_13228607 | 9.68 |
ENSRNOT00000042010
ENSRNOT00000088214 |
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr1_-_222167447 | 9.64 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr13_-_56877611 | 9.59 |
ENSRNOT00000079040
ENSRNOT00000017195 |
Cfhr1
|
complement factor H-related 1 |
| chr5_+_124300477 | 9.58 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
| chr4_+_25538497 | 9.57 |
ENSRNOT00000009264
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr3_+_113319456 | 9.56 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr12_-_1195591 | 9.44 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chr18_-_55916220 | 9.42 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr18_+_29966245 | 9.38 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_+_248723397 | 9.30 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr6_-_27582985 | 9.27 |
ENSRNOT00000085613
ENSRNOT00000014637 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr1_+_72545596 | 9.20 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr4_+_10295122 | 9.12 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr14_-_108372068 | 9.08 |
ENSRNOT00000088116
ENSRNOT00000091143 ENSRNOT00000085886 |
RGD1305110
|
similar to KIAA1841 protein |
| chr15_+_17834635 | 9.06 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr5_-_78324278 | 9.06 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr14_-_21128505 | 9.01 |
ENSRNOT00000004776
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr3_+_48096954 | 8.66 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr11_-_28527890 | 8.65 |
ENSRNOT00000002138
|
Cldn8
|
claudin 8 |
| chr10_+_70262361 | 8.56 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr5_-_19368431 | 8.52 |
ENSRNOT00000012819
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr10_+_53570989 | 8.30 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr3_+_131351587 | 8.24 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr10_+_14240219 | 8.23 |
ENSRNOT00000020233
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr4_+_30313102 | 8.13 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chrM_+_2740 | 8.11 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr3_+_117422704 | 8.04 |
ENSRNOT00000085647
|
Slc12a1
|
solute carrier family 12 member 1 |
| chr11_-_4397361 | 8.03 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr9_+_4107246 | 7.93 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr18_+_59748444 | 7.90 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr4_-_159399634 | 7.87 |
ENSRNOT00000089193
|
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr16_-_7026540 | 7.84 |
ENSRNOT00000051751
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr4_-_160662974 | 7.82 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chr11_+_64882288 | 7.80 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr4_+_35279063 | 7.79 |
ENSRNOT00000011833
|
Nxph1
|
neurexophilin 1 |
| chr1_-_225283326 | 7.70 |
ENSRNOT00000027342
|
Scgb1a1
|
secretoglobin family 1A member 1 |
| chr18_+_30527705 | 7.69 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr9_+_20951260 | 7.61 |
ENSRNOT00000016906
|
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr3_-_160038078 | 7.55 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr10_-_44746549 | 7.47 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr13_-_97838228 | 7.39 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr11_+_74984613 | 7.37 |
ENSRNOT00000035049
|
Atp13a5
|
ATPase 13A5 |
| chr14_-_21127868 | 7.24 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chrX_-_83151511 | 7.20 |
ENSRNOT00000057378
|
Hdx
|
highly divergent homeobox |
| chrX_+_140175861 | 7.19 |
ENSRNOT00000071222
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr7_-_95310005 | 7.17 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chrX_-_34162827 | 7.15 |
ENSRNOT00000046759
|
RGD1560203
|
similar to ferritin heavy polypeptide-like 17 |
| chr2_-_261337163 | 7.08 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr20_-_31598118 | 6.91 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr6_+_101603319 | 6.88 |
ENSRNOT00000030470
|
Gphn
|
gephyrin |
| chr13_+_78979321 | 6.84 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr1_-_101773508 | 6.77 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr2_-_96668222 | 6.54 |
ENSRNOT00000016567
|
Pkia
|
cAMP-dependent protein kinase inhibitor alpha |
| chr1_+_264741911 | 6.51 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chrX_-_15428518 | 6.48 |
ENSRNOT00000075774
|
LOC108348172
|
proSAAS |
| chr3_+_129018592 | 6.35 |
ENSRNOT00000007274
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr10_-_27179254 | 6.33 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chrM_+_14136 | 6.24 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr16_-_74710704 | 5.97 |
ENSRNOT00000016859
|
Thsd1
|
thrombospondin type 1 domain containing 1 |
| chrX_-_15327684 | 5.85 |
ENSRNOT00000009794
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr3_-_168018410 | 5.80 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr4_+_24222500 | 5.71 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr7_+_142776252 | 5.67 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr6_+_27535020 | 5.65 |
ENSRNOT00000076512
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr16_-_6404578 | 5.54 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr8_+_71216178 | 5.49 |
ENSRNOT00000021372
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr2_+_58462588 | 5.48 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr17_-_21739408 | 5.44 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr14_-_66978499 | 5.35 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr10_+_78416946 | 5.33 |
ENSRNOT00000042211
|
AABR07030263.1
|
|
| chr14_+_23089699 | 5.28 |
ENSRNOT00000036948
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr1_+_164380577 | 5.21 |
ENSRNOT00000055321
|
Gdpd5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr10_-_67478848 | 5.18 |
ENSRNOT00000005325
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr5_-_60655985 | 5.18 |
ENSRNOT00000093103
|
Fbxo10
|
F-box protein 10 |
| chr2_-_100249811 | 5.16 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_94718402 | 5.15 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr3_-_71845232 | 5.13 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr1_-_228538074 | 5.11 |
ENSRNOT00000028604
|
Olr324
|
olfactory receptor 324 |
| chr1_+_30681681 | 5.07 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr3_-_76496283 | 5.01 |
ENSRNOT00000044305
|
Olr611
|
olfactory receptor 611 |
| chr3_+_95233874 | 5.00 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr7_+_58814805 | 4.92 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr1_-_101500850 | 4.92 |
ENSRNOT00000028390
|
Nucb1
|
nucleobindin 1 |
| chr1_+_234252757 | 4.80 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr20_-_2851396 | 4.78 |
ENSRNOT00000081234
|
Btnl8
|
butyrophilin-like 8 |
| chr2_+_260444227 | 4.77 |
ENSRNOT00000064249
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr1_+_172792874 | 4.76 |
ENSRNOT00000077837
|
Olr270
|
olfactory receptor 270 |
| chr7_+_15785410 | 4.76 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chrX_+_83926513 | 4.67 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr8_+_50139997 | 4.62 |
ENSRNOT00000022796
|
Bace1
|
beta-secretase 1 |
| chr1_+_178636544 | 4.62 |
ENSRNOT00000092099
|
Spon1
|
spondin 1 |
| chr7_-_8060373 | 4.40 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr10_+_103956019 | 4.37 |
ENSRNOT00000045875
|
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr9_-_113246545 | 4.27 |
ENSRNOT00000034759
|
Tmem232
|
transmembrane protein 232 |
| chr2_+_186980793 | 4.18 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr10_-_74298599 | 4.13 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr13_-_1946508 | 4.01 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr10_+_92769031 | 3.98 |
ENSRNOT00000090759
|
AABR07030543.1
|
|
| chr10_-_98469799 | 3.90 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr6_+_122603269 | 3.89 |
ENSRNOT00000005297
|
Spata7
|
spermatogenesis associated 7 |
| chr20_+_44060731 | 3.88 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr15_-_77736892 | 3.87 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr2_+_208749996 | 3.82 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr1_-_267203986 | 3.75 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr9_-_65693822 | 3.75 |
ENSRNOT00000038431
|
Als2cr12
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 12 |
| chr4_-_82295470 | 3.74 |
ENSRNOT00000091073
|
Hoxa10
|
homeobox A10 |
| chr2_+_144861455 | 3.68 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr3_+_55960067 | 3.68 |
ENSRNOT00000010216
|
Ppig
|
peptidylprolyl isomerase G |
| chr10_-_93675991 | 3.66 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr15_+_56757315 | 3.61 |
ENSRNOT00000078013
|
Esd
|
esterase D |
| chrX_+_43881246 | 3.60 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr13_-_69780956 | 3.56 |
ENSRNOT00000087713
ENSRNOT00000065521 |
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr1_+_154606490 | 3.49 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr2_+_241909832 | 3.49 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_-_95502775 | 3.43 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr1_-_156327352 | 3.41 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr10_+_84135116 | 3.40 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr4_-_82209933 | 3.38 |
ENSRNOT00000091106
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr9_-_15646381 | 3.38 |
ENSRNOT00000040204
|
Mrps10
|
mitochondrial ribosomal protein S10 |
| chr4_+_88423412 | 3.36 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr11_+_69739384 | 3.34 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_+_69415057 | 3.32 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr7_+_36826360 | 3.32 |
ENSRNOT00000029783
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr3_-_73543088 | 3.28 |
ENSRNOT00000012912
|
Olr485
|
olfactory receptor 485 |
| chr20_-_2817642 | 3.27 |
ENSRNOT00000086942
|
LOC100911938
|
butyrophilin subfamily 3 member A3-like |
| chr1_-_164307084 | 3.22 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr3_-_45498327 | 3.21 |
ENSRNOT00000090213
|
AABR07052296.1
|
|
| chr2_+_208750356 | 3.20 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr10_+_13214130 | 3.19 |
ENSRNOT00000073065
|
Prss21
|
protease, serine, 21 |
| chr11_-_87924816 | 3.18 |
ENSRNOT00000031819
|
Serpind1
|
serpin family D member 1 |
| chr5_+_118574801 | 3.18 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_+_168196862 | 3.08 |
ENSRNOT00000021006
|
Olr74
|
olfactory receptor 74 |
| chr7_-_9592000 | 3.07 |
ENSRNOT00000011188
|
Olr1071
|
olfactory receptor 1071 |
| chr10_+_59923201 | 3.03 |
ENSRNOT00000073603
ENSRNOT00000045573 |
Olr1498
|
olfactory receptor 1498 |
| chr11_+_44072043 | 3.01 |
ENSRNOT00000033716
|
LOC680520
|
similar to CG4349-PA |
| chr4_-_119756765 | 2.98 |
ENSRNOT00000013884
|
Cnbp
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr4_+_118814284 | 2.96 |
ENSRNOT00000024884
|
Nfu1
|
NFU1 iron-sulfur cluster scaffold |
| chr7_-_98197087 | 2.93 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr2_+_60180215 | 2.91 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr4_-_168382837 | 2.90 |
ENSRNOT00000029510
|
Mansc1
|
MANSC domain containing 1 |
| chr17_-_21484456 | 2.87 |
ENSRNOT00000038180
|
Sycp2l
|
synaptonemal complex protein 2-like |
| chr2_+_155555840 | 2.80 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr1_-_170175350 | 2.77 |
ENSRNOT00000023349
|
Olr201
|
olfactory receptor 201 |
| chr3_+_117421604 | 2.75 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr18_+_66741144 | 2.60 |
ENSRNOT00000021161
|
AABR07032457.1
|
|
| chr7_+_142776580 | 2.56 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr8_-_128416650 | 2.48 |
ENSRNOT00000046864
|
Scn10a
|
sodium voltage-gated channel alpha subunit 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.2 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 5.0 | 20.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 4.7 | 14.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 4.6 | 18.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 4.4 | 26.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 3.9 | 11.6 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 3.6 | 10.8 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.6 | 10.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 3.3 | 9.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 3.3 | 9.9 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 3.3 | 9.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 3.2 | 9.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 3.2 | 12.7 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 3.1 | 9.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 2.6 | 7.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 2.5 | 10.0 | GO:0051958 | methotrexate transport(GO:0051958) |
| 2.4 | 9.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 2.2 | 10.8 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.0 | 14.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.9 | 9.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 1.9 | 7.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 1.9 | 7.5 | GO:0009597 | detection of virus(GO:0009597) |
| 1.8 | 12.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 1.8 | 5.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 1.8 | 11.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.8 | 5.3 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 1.7 | 17.3 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 1.6 | 16.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.5 | 10.2 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 1.4 | 7.0 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.4 | 9.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.3 | 14.4 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 1.3 | 10.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.2 | 31.7 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 1.1 | 13.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 10.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 1.1 | 5.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.0 | 10.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.0 | 6.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.0 | 12.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.0 | 9.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 5.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.9 | 11.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.9 | 3.5 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.8 | 4.0 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.8 | 10.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.7 | 8.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.7 | 8.5 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.7 | 5.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.7 | 9.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.7 | 9.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 1.3 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 8.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.6 | 3.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.6 | 16.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.6 | 3.6 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.6 | 6.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 19.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.5 | 16.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.5 | 4.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.5 | 7.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 3.9 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.5 | 8.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.5 | 6.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 12.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 9.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 1.2 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.4 | 9.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.4 | 3.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.4 | 18.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.4 | 1.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.4 | 2.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.4 | 5.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 5.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 3.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 2.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 17.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.3 | 1.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 5.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.3 | 10.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 7.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 11.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 1.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 6.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 1.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 | 4.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 18.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 1.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 1.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 2.0 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.6 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.2 | 1.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 3.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 3.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 1.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 8.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 3.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 9.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 9.6 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 4.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 3.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 2.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 15.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 7.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 3.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 3.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 8.7 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 11.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 15.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 6.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 2.0 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 51.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.4 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 17.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 5.2 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.5 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 6.8 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 6.6 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.3 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 23.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 3.1 | 9.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 2.0 | 9.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.6 | 4.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 1.6 | 12.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.4 | 8.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.4 | 8.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.4 | 9.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.3 | 11.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.1 | 10.7 | GO:0034464 | BBSome(GO:0034464) |
| 1.0 | 8.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.0 | 7.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.0 | 18.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.9 | 11.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.9 | 16.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.8 | 3.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.8 | 6.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.6 | 12.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 25.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.6 | 3.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 9.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.5 | 1.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 14.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.4 | 21.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 8.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 1.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 27.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.4 | 14.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 26.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 36.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 16.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 12.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.3 | 5.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 7.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 8.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 23.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 22.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 11.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 6.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.2 | 7.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 3.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 8.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 13.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 10.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 23.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 14.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 9.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 5.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 15.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 6.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 7.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 7.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 10.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 8.2 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 5.2 | 26.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 3.9 | 11.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 3.6 | 10.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 3.6 | 10.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 3.5 | 10.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 3.3 | 9.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 3.1 | 9.3 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 2.8 | 14.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 2.6 | 7.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.5 | 10.0 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 2.5 | 9.9 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 2.5 | 7.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 2.4 | 9.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 2.3 | 6.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 2.1 | 12.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.9 | 11.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.8 | 5.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.7 | 5.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.7 | 6.9 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 1.7 | 5.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 1.6 | 19.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.6 | 31.7 | GO:0051184 | cofactor transporter activity(GO:0051184) modified amino acid transmembrane transporter activity(GO:0072349) |
| 1.6 | 14.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.5 | 7.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.4 | 31.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.4 | 12.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.4 | 11.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.4 | 8.2 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 1.4 | 9.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.4 | 5.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 6.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.3 | 4.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 1.3 | 5.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 1.3 | 7.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.2 | 23.2 | GO:0001848 | complement binding(GO:0001848) |
| 1.2 | 7.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 1.2 | 7.0 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.1 | 4.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.0 | 2.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.9 | 5.5 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.9 | 3.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 5.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.8 | 13.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 10.3 | GO:0098918 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.7 | 3.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.7 | 7.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.7 | 13.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.7 | 2.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.7 | 16.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.6 | 11.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 16.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.6 | 11.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 6.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 10.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 12.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.5 | 6.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 46.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 1.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.4 | 12.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 2.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 3.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 7.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 11.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 14.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.3 | 8.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 8.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 5.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 8.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 10.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 5.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 16.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 4.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 4.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 12.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 2.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 5.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 4.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 14.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 12.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 11.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 3.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 9.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 17.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 4.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 9.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 23.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 7.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 11.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 6.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 3.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 8.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 3.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 26.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 4.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 5.0 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 26.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.9 | 11.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.9 | 16.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.8 | 42.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.6 | 23.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.5 | 20.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 8.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 9.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 9.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 8.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 17.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 23.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 3.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.7 | 23.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 20.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.9 | 26.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.7 | 11.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.7 | 11.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 13.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.7 | 9.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 14.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 8.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.6 | 12.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.5 | 6.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 19.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.5 | 9.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 8.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 11.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 6.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 11.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 8.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 5.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 16.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 4.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 5.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 10.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 25.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 27.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 5.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 6.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 6.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 6.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 11.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 6.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.1 | 5.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |