Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
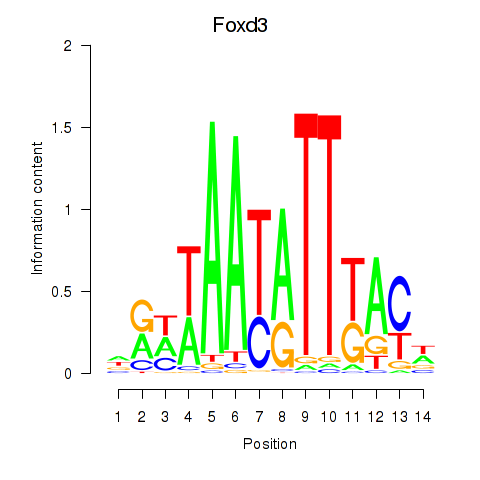
Results for Foxd3
Z-value: 0.62
Transcription factors associated with Foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Foxd3 motif
Sorted Z-values of Foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_127620296 | 33.89 |
ENSRNOT00000012577
|
Serpina1
|
serpin family A member 1 |
| chr9_+_95501778 | 30.02 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr7_-_143523457 | 26.31 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr16_+_50181316 | 24.41 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr16_+_18736154 | 22.16 |
ENSRNOT00000015723
|
Mbl1
|
mannose-binding lectin (protein A) 1 |
| chr7_+_34326087 | 21.52 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr13_+_27465930 | 20.93 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr13_+_91080341 | 19.90 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr1_-_170431073 | 18.16 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr2_+_235264219 | 17.77 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr6_-_127508452 | 16.77 |
ENSRNOT00000073709
|
LOC100909524
|
protein Z-dependent protease inhibitor-like |
| chr3_+_149668102 | 15.12 |
ENSRNOT00000055342
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr8_-_21901829 | 15.00 |
ENSRNOT00000027963
|
Angptl6
|
angiopoietin-like 6 |
| chr15_-_44627765 | 10.23 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chrX_+_40460047 | 9.04 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_+_176510696 | 8.50 |
ENSRNOT00000016614
|
Iapp
|
islet amyloid polypeptide |
| chr4_-_28437676 | 6.41 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr8_-_97647072 | 5.39 |
ENSRNOT00000030645
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr16_-_48692476 | 5.19 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr4_-_163403653 | 5.04 |
ENSRNOT00000088151
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr4_+_66670618 | 4.84 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr10_+_64174931 | 4.72 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr6_-_128434183 | 4.47 |
ENSRNOT00000014405
|
Dicer1
|
dicer 1 ribonuclease III |
| chr2_-_186232292 | 2.56 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr3_-_12155098 | 2.55 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chrX_-_71169865 | 2.00 |
ENSRNOT00000050415
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr1_-_164977633 | 1.81 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr1_+_265809754 | 1.70 |
ENSRNOT00000025221
|
Pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr5_+_60850852 | 1.59 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr7_+_51794173 | 1.28 |
ENSRNOT00000043774
|
Otogl
|
otogelin-like |
| chr4_-_168517177 | 1.25 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr1_-_124803363 | 1.02 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr13_+_53351717 | 0.91 |
ENSRNOT00000012038
|
Kif14
|
kinesin family member 14 |
| chr15_+_62406873 | 0.83 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr17_-_34945317 | 0.74 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr15_-_32888095 | 0.73 |
ENSRNOT00000012233
|
Dad1
|
defender against cell death 1 |
| chr1_+_259958310 | 0.71 |
ENSRNOT00000019751
|
LOC103689954
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr10_+_11338306 | 0.55 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr12_-_12782139 | 0.51 |
ENSRNOT00000001392
ENSRNOT00000079836 |
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr10_+_10889488 | 0.47 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr3_-_73755510 | 0.40 |
ENSRNOT00000083628
|
Olr502
|
olfactory receptor 502 |
| chr3_-_163847671 | 0.38 |
ENSRNOT00000076833
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr14_+_46001849 | 0.31 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr5_-_136112344 | 0.30 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr5_-_83648044 | 0.09 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr8_-_19527513 | 0.05 |
ENSRNOT00000008594
|
Olr1147
|
olfactory receptor 1147 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 33.9 | GO:0033986 | response to methanol(GO:0033986) |
| 7.2 | 21.5 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 4.5 | 18.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 3.5 | 24.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 3.4 | 10.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.3 | 19.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 3.0 | 9.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 1.7 | 22.2 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 1.7 | 5.0 | GO:2000502 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 1.6 | 4.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.5 | 15.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 1.5 | 4.5 | GO:0032290 | zygote asymmetric cell division(GO:0010070) targeting of mRNA for destruction involved in RNA interference(GO:0030423) peripheral nervous system myelin formation(GO:0032290) |
| 0.6 | 17.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.5 | 4.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.5 | 8.5 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.3 | 0.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 27.7 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.2 | 2.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 23.9 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.1 | 2.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 35.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.1 | 1.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.8 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 1.8 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 5.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.4 | 26.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 22.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 18.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 15.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 118.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 15.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 7.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 5.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 32.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 12.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 21.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 2.8 | 19.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.0 | 24.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 2.0 | 18.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.1 | 4.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 1.1 | 22.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 69.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 17.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 2.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.3 | 1.6 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 5.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 4.8 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 10.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 5.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 25.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 7.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 10.9 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 4.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 33.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 19.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 18.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 20.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 15.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.3 | 17.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.2 | 24.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 33.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 8.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 5.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 4.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 4.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 21.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 5.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 10.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |