Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
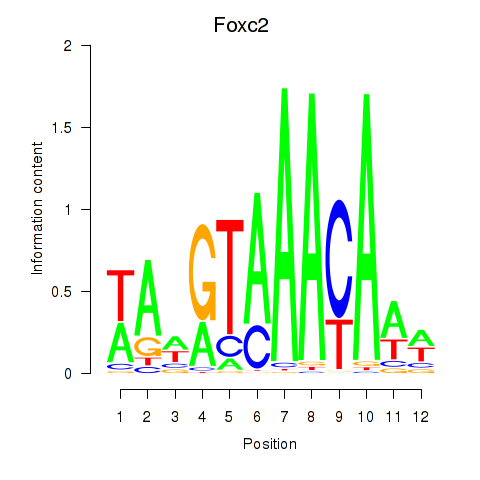
Results for Foxc2
Z-value: 1.19
Transcription factors associated with Foxc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc2
|
ENSRNOG00000047446 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc2 | rn6_v1_chr19_+_53044379_53044379 | -0.09 | 1.1e-01 | Click! |
Activity profile of Foxc2 motif
Sorted Z-values of Foxc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_225283326 | 133.04 |
ENSRNOT00000027342
|
Scgb1a1
|
secretoglobin family 1A member 1 |
| chr3_+_149624712 | 80.19 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr20_-_31598118 | 71.24 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr2_+_208749996 | 46.39 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr2_+_208750356 | 46.30 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr7_-_143523457 | 40.66 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr4_-_115239723 | 40.17 |
ENSRNOT00000042699
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr20_-_31597830 | 40.00 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr18_+_68408890 | 31.39 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr18_+_3887419 | 27.74 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr9_-_55256340 | 26.90 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr7_+_58814805 | 24.21 |
ENSRNOT00000005909
|
Tspan8
|
tetraspanin 8 |
| chr8_+_58431407 | 22.83 |
ENSRNOT00000011974
|
Sln
|
sarcolipin |
| chr16_+_50181316 | 21.55 |
ENSRNOT00000077662
|
F11
|
coagulation factor XI |
| chr15_-_95514259 | 21.43 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr4_-_176528110 | 20.94 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr13_-_44540516 | 19.99 |
ENSRNOT00000005244
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr3_+_162357356 | 15.27 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr2_-_3124543 | 12.51 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr16_-_23156962 | 12.40 |
ENSRNOT00000018543
|
Sh2d4a
|
SH2 domain containing 4A |
| chr2_-_233743866 | 11.83 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr4_+_21462779 | 11.71 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr4_-_15505362 | 11.55 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr7_+_142877086 | 11.40 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr7_-_107392972 | 11.02 |
ENSRNOT00000093425
|
Tmem71
|
transmembrane protein 71 |
| chr11_-_45124423 | 10.39 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr20_+_29655226 | 9.57 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr6_-_78549669 | 9.50 |
ENSRNOT00000009940
|
Foxa1
|
forkhead box A1 |
| chr1_-_267203986 | 9.44 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_-_156327352 | 8.23 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr18_-_70924708 | 8.20 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr1_+_154606490 | 8.03 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr9_+_51298426 | 7.85 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_-_172361779 | 7.66 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr9_+_118849302 | 7.38 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr10_-_103687425 | 7.38 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr4_+_139670092 | 7.31 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr3_-_63535991 | 6.52 |
ENSRNOT00000015722
|
Fkbp7
|
FK506 binding protein 7 |
| chr4_+_62221011 | 6.20 |
ENSRNOT00000041264
|
Cald1
|
caldesmon 1 |
| chr1_+_227592635 | 6.16 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr3_-_160038078 | 6.15 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr1_-_89269930 | 6.02 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr5_+_124690214 | 5.84 |
ENSRNOT00000011237
|
Plpp3
|
phospholipid phosphatase 3 |
| chr2_+_145174876 | 5.71 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr4_+_14070553 | 5.70 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr2_-_30634243 | 5.56 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr4_+_62220736 | 4.88 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr2_-_5577369 | 4.55 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr8_-_61595032 | 4.52 |
ENSRNOT00000023428
|
Snx33
|
sorting nexin 33 |
| chr9_-_82345262 | 3.61 |
ENSRNOT00000024975
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr2_+_143475323 | 2.84 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr20_+_46429222 | 2.63 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr8_-_55402711 | 2.59 |
ENSRNOT00000091732
|
Sik2
|
salt-inducible kinase 2 |
| chr2_-_258997138 | 2.58 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr4_+_41364441 | 2.53 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr18_-_5314511 | 2.28 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr10_-_95434688 | 2.21 |
ENSRNOT00000074809
|
AABR07030603.1
|
|
| chr5_+_103251986 | 1.89 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr15_+_24942218 | 1.75 |
ENSRNOT00000016629
|
Peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_-_11585078 | 1.25 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr13_+_78979321 | 0.99 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr18_-_69671199 | 0.97 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr16_+_27399467 | 0.79 |
ENSRNOT00000065642
|
Tll1
|
tolloid-like 1 |
| chr10_+_105393072 | 0.75 |
ENSRNOT00000013359
|
Ubald2
|
UBA-like domain containing 2 |
| chr6_-_51257625 | 0.68 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
| chr17_-_21739408 | 0.68 |
ENSRNOT00000060335
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr4_+_62380914 | 0.68 |
ENSRNOT00000029845
|
Tmem140
|
transmembrane protein 140 |
| chr7_-_114848414 | 0.68 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr3_-_60023913 | 0.44 |
ENSRNOT00000064662
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr14_+_20266891 | 0.36 |
ENSRNOT00000004174
|
Gc
|
group specific component |
| chr5_+_50381244 | 0.23 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr13_-_78521911 | 0.11 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr16_+_2634603 | 0.02 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.3 | 133.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 18.5 | 92.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 16.0 | 80.2 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 8.0 | 40.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 4.3 | 21.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 3.3 | 22.8 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 3.1 | 111.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 3.1 | 21.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 2.9 | 11.6 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 2.7 | 8.2 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 2.5 | 7.4 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 2.4 | 11.8 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.9 | 5.8 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 1.9 | 9.5 | GO:0061144 | prostate gland stromal morphogenesis(GO:0060741) alveolar secondary septum development(GO:0061144) |
| 1.7 | 15.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 1.6 | 9.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.5 | 6.2 | GO:0009597 | detection of virus(GO:0009597) |
| 1.5 | 6.0 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 1.5 | 27.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.2 | 9.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 11.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.9 | 5.7 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.9 | 7.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.8 | 9.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 24.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.5 | 4.5 | GO:0036089 | cleavage furrow formation(GO:0036089) macropinocytosis(GO:0044351) |
| 0.5 | 1.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.4 | 2.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.4 | 7.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 4.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 2.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 1.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 40.0 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 7.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 6.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.8 | GO:0014051 | manganese ion transport(GO:0006828) gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 10.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 5.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 11.1 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 9.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 3.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 1.4 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 27.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 2.7 | 40.2 | GO:0032982 | myosin filament(GO:0032982) |
| 1.8 | 11.1 | GO:0030478 | actin cap(GO:0030478) |
| 1.4 | 108.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 1.1 | 133.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 1.1 | 5.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 11.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.8 | 89.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.7 | 11.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.7 | 22.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 26.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 22.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 29.7 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 9.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 9.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 96.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 12.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 29.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.6 | 133.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 15.4 | 92.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 3.9 | 11.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.6 | 20.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.9 | 5.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.8 | 7.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.8 | 21.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.4 | 8.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.1 | 5.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.0 | 11.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.9 | 9.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.6 | 11.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 110.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.6 | 4.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 7.4 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.5 | 11.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 6.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 6.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 26.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 9.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.7 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 24.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.8 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 11.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 86.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.1 | 1.0 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 17.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 2.6 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 10.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 30.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 22.3 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.0 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 19.1 | GO:0005524 | ATP binding(GO:0005524) |
| 0.0 | 5.1 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 3.6 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 141.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 2.5 | 122.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 1.7 | 27.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 40.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 9.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 20.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.9 | 112.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 1.9 | 51.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.0 | 21.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 11.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 11.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 27.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 2.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 1.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 5.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 6.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |