Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
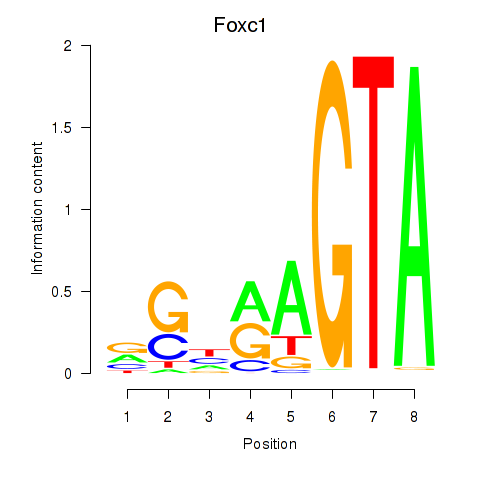
Results for Foxc1
Z-value: 0.86
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSRNOG00000017800 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | rn6_v1_chr17_-_33951484_33951484 | 0.11 | 5.3e-02 | Click! |
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_215610368 | 31.41 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr10_+_53781239 | 30.28 |
ENSRNOT00000082871
|
Myh2
|
myosin heavy chain 2 |
| chr1_+_215609645 | 24.95 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr10_+_53713938 | 22.43 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr2_+_60131776 | 21.82 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr16_+_25773602 | 18.91 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr10_+_59725398 | 18.33 |
ENSRNOT00000026156
|
P2rx5
|
purinergic receptor P2X 5 |
| chr7_-_113937941 | 17.86 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr11_-_72109964 | 17.82 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr5_+_115649046 | 16.12 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_-_102289837 | 15.39 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr1_-_253000760 | 14.10 |
ENSRNOT00000030024
|
Slc16a12
|
solute carrier family 16, member 12 |
| chr8_+_100260049 | 14.09 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr16_+_16949232 | 13.83 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr13_-_103080920 | 13.71 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr16_+_26906716 | 13.02 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chr7_-_49250953 | 12.52 |
ENSRNOT00000066975
ENSRNOT00000082141 |
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr2_+_95077577 | 12.45 |
ENSRNOT00000046368
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr1_+_193537137 | 12.01 |
ENSRNOT00000029967
|
AABR07005667.1
|
|
| chr6_+_64297888 | 11.88 |
ENSRNOT00000050222
ENSRNOT00000083088 ENSRNOT00000093147 |
Nrcam
|
neuronal cell adhesion molecule |
| chr1_-_82452281 | 11.83 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr12_-_5773036 | 11.74 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr3_-_160301552 | 11.42 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr17_-_84614228 | 11.36 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr10_+_56662242 | 11.06 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr14_+_37116492 | 10.81 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr10_+_56662561 | 10.47 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr1_+_101625790 | 10.45 |
ENSRNOT00000029220
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr18_-_71614980 | 9.92 |
ENSRNOT00000032563
|
LOC102548286
|
peroxisomal biogenesis factor 19-like |
| chr8_-_109560747 | 9.83 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_-_64952687 | 9.56 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr20_-_45126062 | 9.40 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chrX_+_142344878 | 8.78 |
ENSRNOT00000056621
|
LOC108349258
|
high mobility group protein B4-like |
| chr10_-_84698886 | 8.50 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr13_+_56262190 | 8.33 |
ENSRNOT00000032908
|
AABR07021086.1
|
|
| chr14_-_19132208 | 8.27 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr13_+_52667969 | 8.25 |
ENSRNOT00000084986
ENSRNOT00000050284 |
Tnnt2
|
troponin T2, cardiac type |
| chr9_+_52023295 | 8.17 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr3_+_9643047 | 8.16 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr13_-_111972603 | 8.08 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_-_258877045 | 7.98 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr3_+_60024013 | 7.93 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr1_+_150067242 | 7.72 |
ENSRNOT00000073824
|
Olr23
|
olfactory receptor 23 |
| chr1_-_251379593 | 7.55 |
ENSRNOT00000014684
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr10_-_98544447 | 7.18 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr14_-_14689554 | 6.90 |
ENSRNOT00000002814
|
Fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr2_+_84678948 | 6.86 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr18_-_35817117 | 6.29 |
ENSRNOT00000015172
ENSRNOT00000085714 |
Mcc
|
mutated in colorectal cancers |
| chr19_+_50246402 | 6.29 |
ENSRNOT00000018795
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_-_96032722 | 6.20 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr3_-_75089064 | 6.17 |
ENSRNOT00000046167
|
Olr542
|
olfactory receptor 542 |
| chr8_-_39266959 | 5.87 |
ENSRNOT00000046590
|
Ei24
|
EI24, autophagy associated transmembrane protein |
| chr7_-_144269486 | 5.59 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr3_-_91195981 | 5.47 |
ENSRNOT00000056935
|
RGD1309730
|
similar to RIKEN cDNA B230118H07 |
| chr6_+_64252970 | 5.45 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr10_+_13196946 | 5.37 |
ENSRNOT00000071525
|
Prss32
|
protease, serine, 32 |
| chr16_-_79973735 | 5.25 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr2_-_258997138 | 5.11 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr11_+_28692708 | 4.93 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr3_+_122544788 | 4.83 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr4_+_72292261 | 4.64 |
ENSRNOT00000078723
|
LOC103690255
|
olfactory receptor-like protein OLF3 |
| chr16_-_7758189 | 4.61 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr3_-_77710180 | 4.57 |
ENSRNOT00000087015
|
Olr671
|
olfactory receptor 671 |
| chr15_-_33263659 | 4.53 |
ENSRNOT00000018005
|
Psmb5
|
proteasome subunit beta 5 |
| chr10_-_78690857 | 4.50 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr4_+_158243086 | 4.47 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr20_-_9855443 | 4.41 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chrX_+_75382598 | 4.30 |
ENSRNOT00000033494
|
Uprt
|
uracil phosphoribosyltransferase homolog |
| chr2_-_235232530 | 4.23 |
ENSRNOT00000093386
ENSRNOT00000079943 |
Lrit3
|
leucine-rich repeat, Ig-like and transmembrane domains 3 |
| chr11_-_34598102 | 4.23 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_-_197981665 | 4.22 |
ENSRNOT00000083086
|
Mrps21
|
mitochondrial ribosomal protein S21 |
| chrX_+_71342775 | 4.14 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr8_+_22625874 | 4.11 |
ENSRNOT00000012269
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr3_+_114253637 | 4.03 |
ENSRNOT00000046843
|
Duox1
|
dual oxidase 1 |
| chr1_+_149963547 | 3.95 |
ENSRNOT00000042245
|
Olr20
|
olfactory receptor 20 |
| chr10_+_88992487 | 3.93 |
ENSRNOT00000027061
|
Coasy
|
Coenzyme A synthase |
| chr8_+_57936650 | 3.79 |
ENSRNOT00000089686
|
Exph5
|
exophilin 5 |
| chr3_-_21027947 | 3.79 |
ENSRNOT00000051973
|
Olr421
|
olfactory receptor 421 |
| chr2_+_112914375 | 3.64 |
ENSRNOT00000092737
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr1_+_70480941 | 3.61 |
ENSRNOT00000072404
|
Olr6
|
olfactory receptor 6 |
| chr12_+_19513100 | 3.58 |
ENSRNOT00000030161
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr5_-_105579959 | 3.48 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr1_+_230628996 | 3.46 |
ENSRNOT00000017604
|
Olr376
|
olfactory receptor 376 |
| chr1_-_226732736 | 3.33 |
ENSRNOT00000072343
|
LOC108348129
|
pepsin F-like |
| chr3_+_128756799 | 3.27 |
ENSRNOT00000049855
ENSRNOT00000042853 |
Plcb4
|
phospholipase C, beta 4 |
| chr15_-_32925673 | 3.26 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr13_-_86671515 | 3.22 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr1_-_70485888 | 3.22 |
ENSRNOT00000020514
|
Olr7
|
olfactory receptor 7 |
| chr2_+_77084222 | 3.20 |
ENSRNOT00000087171
|
Gm9733
|
predicted gene 9733 |
| chr10_-_39622973 | 3.20 |
ENSRNOT00000031762
|
Il3
|
interleukin 3 |
| chr12_+_19512591 | 3.07 |
ENSRNOT00000060024
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr8_-_36876931 | 3.01 |
ENSRNOT00000073753
|
Pate1
|
prostate and testis expressed 1 |
| chr1_+_150130084 | 2.95 |
ENSRNOT00000041243
|
Olr25
|
olfactory receptor 25 |
| chr5_+_76092287 | 2.90 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr18_+_56379890 | 2.89 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr9_-_26734763 | 2.75 |
ENSRNOT00000082814
|
AABR07067024.1
|
|
| chr7_+_121480723 | 2.74 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr3_-_79165698 | 2.70 |
ENSRNOT00000032617
|
Olr744
|
olfactory receptor 744 |
| chr1_-_172322795 | 2.69 |
ENSRNOT00000075318
|
RGD1562400
|
similar to olfactory receptor MOR204-14 |
| chr5_+_137564607 | 2.66 |
ENSRNOT00000075742
|
LOC100912529
|
olfactory receptor 2B6-like |
| chr8_+_43355060 | 2.64 |
ENSRNOT00000043206
|
Olr1309
|
olfactory receptor 1309 |
| chr1_+_44311513 | 2.60 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_+_87354042 | 2.57 |
ENSRNOT00000068178
|
LOC100911527
|
interferon alpha-12-like |
| chr15_-_29993639 | 2.55 |
ENSRNOT00000071822
|
AABR07017677.1
|
|
| chr5_+_136406130 | 2.50 |
ENSRNOT00000093099
|
Eri3
|
ERI1 exoribonuclease family member 3 |
| chr9_-_71798265 | 2.50 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chr12_-_2568382 | 2.43 |
ENSRNOT00000035142
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr1_+_258074860 | 2.38 |
ENSRNOT00000054729
|
Cyp2c24
|
cytochrome P450, family 2, subfamily c, polypeptide 24 |
| chr3_+_43255567 | 2.19 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr1_-_261669584 | 2.19 |
ENSRNOT00000020568
ENSRNOT00000076555 |
Crtac1
|
cartilage acidic protein 1 |
| chr4_+_72221550 | 2.11 |
ENSRNOT00000078533
|
Olr806
|
olfactory receptor 806 |
| chr7_+_13996966 | 2.05 |
ENSRNOT00000009615
|
Olr1087
|
olfactory receptor 1087 |
| chr5_+_118574801 | 2.04 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_+_27476375 | 1.84 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr10_-_12236107 | 1.83 |
ENSRNOT00000049390
|
Olr1381
|
olfactory receptor 1381 |
| chr5_+_15043955 | 1.80 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr10_-_12370656 | 1.80 |
ENSRNOT00000060995
|
Olr1362
|
olfactory receptor 1362 |
| chr10_-_71491743 | 1.77 |
ENSRNOT00000038955
|
LOC102552988
|
uncharacterized LOC102552988 |
| chr3_-_162579201 | 1.76 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr1_+_16819170 | 1.74 |
ENSRNOT00000019734
ENSRNOT00000086756 |
Hbs1l
|
HBS1-like translational GTPase |
| chr18_+_30375798 | 1.73 |
ENSRNOT00000060497
|
Pcdhb2
|
protocadherin beta 2 |
| chr3_-_20914603 | 1.67 |
ENSRNOT00000049651
|
Olr416
|
olfactory receptor 416 |
| chr3_-_76289390 | 1.66 |
ENSRNOT00000051968
|
Olr609
|
olfactory receptor 609 |
| chr1_+_150225373 | 1.65 |
ENSRNOT00000051266
|
Olr30
|
olfactory receptor 30 |
| chr5_+_137775801 | 1.63 |
ENSRNOT00000074907
|
LOC686209
|
similar to olfactory receptor 1340 |
| chr10_+_34402482 | 1.54 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr3_+_176865156 | 1.52 |
ENSRNOT00000019084
|
Zgpat
|
zinc finger CCCH-type and G-patch domain containing |
| chr20_+_1781320 | 1.44 |
ENSRNOT00000040388
|
Olr1737
|
olfactory receptor 1737 |
| chr10_-_38969501 | 1.42 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr7_-_144959746 | 1.39 |
ENSRNOT00000064261
|
Zfp385a
|
zinc finger protein 385A |
| chr12_-_19439977 | 1.36 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr2_-_178616719 | 1.35 |
ENSRNOT00000078610
|
Tmem144
|
transmembrane protein 144 |
| chr8_+_59457018 | 1.34 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr1_-_22480522 | 1.32 |
ENSRNOT00000046379
|
Taar7e
|
trace-amine-associated receptor 7e |
| chr8_-_22625959 | 1.32 |
ENSRNOT00000012138
|
Yipf2
|
Yip1 domain family, member 2 |
| chr17_-_77261731 | 1.27 |
ENSRNOT00000066850
|
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr5_-_78285451 | 1.25 |
ENSRNOT00000019924
|
Rnf183
|
ring finger protein 183 |
| chr10_-_12522230 | 1.23 |
ENSRNOT00000073401
|
Olr1370
|
olfactory receptor 1370 |
| chr14_-_37763712 | 1.21 |
ENSRNOT00000030610
|
Zar1
|
zygote arrest 1 |
| chr17_+_25228437 | 1.17 |
ENSRNOT00000072904
|
AABR07027342.1
|
|
| chr16_+_35573058 | 1.09 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr10_-_12777245 | 1.08 |
ENSRNOT00000041708
|
Olr1380
|
olfactory receptor 1380 |
| chr20_-_795286 | 1.04 |
ENSRNOT00000052186
|
Olr1691
|
olfactory receptor 1691 |
| chr20_+_1322453 | 1.00 |
ENSRNOT00000075472
|
Olr1868
|
olfactory receptor 1868 |
| chr9_-_23454316 | 0.95 |
ENSRNOT00000072826
|
Cyp2ac1
|
cytochrome P450, family 2, subfamily ac, polypeptide 1 |
| chr7_-_117068332 | 0.94 |
ENSRNOT00000082433
|
Fam83h
|
family with sequence similarity 83, member H |
| chr8_+_117282390 | 0.89 |
ENSRNOT00000074772
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr15_-_29680444 | 0.89 |
ENSRNOT00000075008
|
AABR07017658.1
|
|
| chr8_+_43382176 | 0.84 |
ENSRNOT00000047759
|
Olr1311
|
olfactory receptor 1311 |
| chr2_-_248484100 | 0.82 |
ENSRNOT00000040856
|
Gbp3
|
guanylate binding protein 3 |
| chr3_+_78835929 | 0.80 |
ENSRNOT00000008762
|
Olr725
|
olfactory receptor 725 |
| chr5_+_107233230 | 0.74 |
ENSRNOT00000029976
|
Ifne
|
interferon, epsilon |
| chr1_-_44615760 | 0.73 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr1_-_175895510 | 0.68 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr18_+_29951094 | 0.64 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr14_+_71533063 | 0.60 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr15_-_37926715 | 0.59 |
ENSRNOT00000013901
|
Xpo4
|
exportin 4 |
| chr13_-_35668968 | 0.59 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr3_-_112876773 | 0.52 |
ENSRNOT00000015086
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr5_-_50138475 | 0.37 |
ENSRNOT00000011969
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr11_+_44001579 | 0.35 |
ENSRNOT00000002258
|
Gpr15
|
G protein-coupled receptor 15 |
| chr8_+_40410604 | 0.26 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr9_+_42620006 | 0.10 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr12_-_40332612 | 0.08 |
ENSRNOT00000001691
|
Atxn2
|
ataxin 2 |
| chr5_-_154394023 | 0.08 |
ENSRNOT00000079823
|
Rpl11
|
ribosomal protein L11 |
| chrX_-_82699487 | 0.04 |
ENSRNOT00000081625
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr14_-_70160624 | 0.01 |
ENSRNOT00000022589
|
Clrn2
|
clarin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 4.1 | 52.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 3.3 | 9.8 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 3.2 | 9.6 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 2.9 | 11.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 2.7 | 8.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.7 | 8.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 2.7 | 8.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.6 | 13.0 | GO:0030070 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 2.4 | 21.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 2.0 | 8.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.7 | 18.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 1.4 | 5.5 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 1.3 | 4.0 | GO:0042335 | cuticle development(GO:0042335) |
| 1.3 | 11.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.1 | 56.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.1 | 10.8 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.0 | 2.9 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.9 | 2.7 | GO:1990737 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.8 | 11.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.8 | 3.2 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.8 | 4.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.7 | 6.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.7 | 5.9 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.6 | 4.3 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.5 | 2.5 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 5.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.5 | 1.4 | GO:2000422 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.4 | 14.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.4 | 1.8 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 2.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.4 | 3.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 7.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 2.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.3 | 1.4 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 8.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 10.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.3 | 3.3 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.3 | 11.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 4.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 3.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 0.6 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.2 | 5.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 6.9 | GO:0003338 | metanephros morphogenesis(GO:0003338) |
| 0.2 | 1.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 2.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 4.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.2 | 0.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 3.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 3.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.7 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.5 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 3.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 21.5 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.1 | 8.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.1 | 3.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 5.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 5.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 66.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 8.9 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 4.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 10.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 4.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 7.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.9 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 6.6 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 3.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:0072678 | T cell migration(GO:0072678) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 52.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 5.4 | 64.6 | GO:0005861 | troponin complex(GO:0005861) |
| 2.4 | 11.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 2.0 | 16.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.8 | 10.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.0 | 18.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.9 | 2.7 | GO:1990589 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.8 | 13.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 8.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 16.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.5 | 2.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 4.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 10.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.4 | 11.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 2.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 5.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 9.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 5.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 11.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 23.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 8.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 13.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 13.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 6.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 9.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 3.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 8.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 66.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 56.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 7.3 | 21.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 4.1 | 8.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 4.0 | 16.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.7 | 8.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 2.7 | 8.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.0 | 18.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.5 | 4.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.3 | 11.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.3 | 11.8 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 1.2 | 13.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.2 | 5.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.1 | 3.2 | GO:0005135 | interleukin-3 receptor binding(GO:0005135) |
| 1.0 | 2.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.9 | 6.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.9 | 3.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.9 | 4.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.8 | 3.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.7 | 8.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 2.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.7 | 14.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.7 | 4.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.7 | 5.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.6 | 7.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 5.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 4.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.5 | 7.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 4.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 10.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 1.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.3 | 10.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 1.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 3.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 5.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 21.5 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.3 | 3.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 9.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 4.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 8.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 0.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 16.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 3.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 4.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 46.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 10.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 9.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 27.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 10.4 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.1 | 7.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 39.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.4 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 4.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.7 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 3.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 11.1 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 3.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 52.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 21.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 13.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 3.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 8.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 6.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 64.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.4 | 17.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.0 | 21.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.7 | 11.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 13.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 7.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 11.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 5.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 8.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 3.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 2.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 3.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |