Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
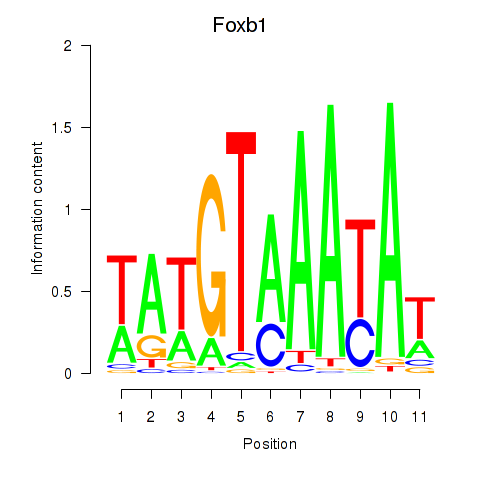
Results for Foxb1
Z-value: 0.40
Transcription factors associated with Foxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxb1
|
ENSRNOG00000010547 | forkhead box B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxb1 | rn6_v1_chr8_-_75995371_75995371 | 0.65 | 1.7e-39 | Click! |
Activity profile of Foxb1 motif
Sorted Z-values of Foxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_2190915 | 11.22 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr11_+_60253608 | 9.11 |
ENSRNOT00000090804
|
LOC685716
|
similar to OX-2 membrane glycoprotein precursor (MRC OX-2 antigen) (CD200 antigen) |
| chr15_-_77736892 | 6.35 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr4_-_40385349 | 6.34 |
ENSRNOT00000039005
|
Gpr85
|
G protein-coupled receptor 85 |
| chr11_-_32858830 | 5.77 |
ENSRNOT00000002313
|
Runx1
|
runt-related transcription factor 1 |
| chr1_-_134870255 | 5.33 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr6_+_42947334 | 5.21 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr5_+_152721940 | 5.14 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr15_-_54906203 | 5.10 |
ENSRNOT00000020186
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr4_-_81241152 | 5.04 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr8_-_80631873 | 4.95 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chrX_+_159703578 | 4.74 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr14_+_24129592 | 4.44 |
ENSRNOT00000040647
|
LOC689899
|
similar to 60S ribosomal protein L23a |
| chr2_+_198388809 | 4.18 |
ENSRNOT00000083087
|
Hist2h2aa3
|
histone cluster 2, H2aa3 |
| chr3_+_94549180 | 4.07 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr2_-_198382190 | 3.94 |
ENSRNOT00000044268
|
Hist2h2aa2
|
histone cluster 2, H2aa2 |
| chr6_+_95826546 | 3.40 |
ENSRNOT00000043587
|
AABR07064810.1
|
|
| chr5_+_164923302 | 3.20 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr13_-_94859390 | 3.12 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr10_-_85825735 | 3.12 |
ENSRNOT00000055379
|
Fbxo47
|
F-box protein 47 |
| chr1_+_220325352 | 3.01 |
ENSRNOT00000027258
|
LOC108348098
|
breast cancer metastasis-suppressor 1 homolog |
| chr19_-_17045349 | 2.96 |
ENSRNOT00000086829
|
Fto
|
FTO, alpha-ketoglutarate dependent dioxygenase |
| chr9_-_15274917 | 2.95 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr13_-_78521911 | 2.91 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr8_-_33661049 | 2.89 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_108660063 | 2.76 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr10_+_68588789 | 2.71 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr2_-_23909016 | 2.68 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr10_-_87351030 | 2.61 |
ENSRNOT00000034889
ENSRNOT00000091152 |
Krt20
|
keratin 20 |
| chr3_+_65816569 | 2.55 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_-_44615760 | 2.47 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr6_+_108745895 | 2.42 |
ENSRNOT00000061786
|
LOC108348062
|
eukaryotic translation initiation factor 3 subunit H |
| chrX_+_78042859 | 2.29 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr5_+_103251986 | 2.27 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr6_-_95061174 | 2.23 |
ENSRNOT00000072262
|
Rtn1
|
reticulon 1 |
| chr1_+_190671696 | 2.16 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr3_+_162357356 | 2.10 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr3_+_151691515 | 2.01 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr3_-_141411170 | 1.93 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr4_+_93959152 | 1.92 |
ENSRNOT00000058437
|
AABR07060795.1
|
|
| chr14_+_42714315 | 1.82 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr10_-_16046033 | 1.79 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_-_11585078 | 1.66 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr9_-_121713091 | 1.52 |
ENSRNOT00000073432
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr17_-_45746753 | 1.37 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr6_-_95890325 | 1.29 |
ENSRNOT00000050217
|
LOC690335
|
similar to 60S ribosomal protein L23a |
| chr2_+_239415046 | 1.22 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr3_-_46153371 | 1.17 |
ENSRNOT00000085885
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr18_+_16146447 | 1.16 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr2_+_143475323 | 1.14 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr20_+_49319307 | 1.05 |
ENSRNOT00000057078
|
Atg5
|
autophagy related 5 |
| chr5_+_157165341 | 0.93 |
ENSRNOT00000087072
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr4_+_139670092 | 0.89 |
ENSRNOT00000008879
|
Lrrn1
|
leucine rich repeat neuronal 1 |
| chr9_+_118849302 | 0.88 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr7_-_107391184 | 0.87 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chrX_-_63203643 | 0.86 |
ENSRNOT00000065194
ENSRNOT00000076974 |
Zfx
|
zinc finger protein X-linked |
| chrX_+_78991223 | 0.76 |
ENSRNOT00000031200
|
Fam46d
|
family with sequence similarity 46, member D |
| chr10_+_54156649 | 0.75 |
ENSRNOT00000074718
|
Gas7
|
growth arrest specific 7 |
| chr3_+_73988945 | 0.73 |
ENSRNOT00000050404
|
Olr515
|
olfactory receptor 515 |
| chr9_-_86103158 | 0.40 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr1_-_89269930 | 0.37 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr3_-_431933 | 0.28 |
ENSRNOT00000033653
|
Spopl
|
speckle type BTB/POZ protein like |
| chr3_-_77525518 | 0.26 |
ENSRNOT00000049513
|
Olr663
|
olfactory receptor 663 |
| chr14_+_35683442 | 0.12 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr3_+_74058255 | 0.11 |
ENSRNOT00000046947
|
Olr517
|
olfactory receptor 517 |
| chr1_-_73961432 | 0.09 |
ENSRNOT00000035435
|
Nlrp4b
|
NLR family, pyrin domain containing 4B |
| chr3_-_73779982 | 0.08 |
ENSRNOT00000079710
|
Olr505
|
olfactory receptor 505 |
| chr3_-_74371385 | 0.08 |
ENSRNOT00000041668
|
Olr518
|
olfactory receptor 518 |
| chr1_+_168830222 | 0.07 |
ENSRNOT00000050333
|
Olr122
|
olfactory receptor 122 |
| chr6_-_51257625 | 0.01 |
ENSRNOT00000012004
|
Hbp1
|
HMG-box transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.0 | 5.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.0 | 3.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.9 | 1.8 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.8 | 5.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.8 | 11.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.6 | 1.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 4.7 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.6 | 3.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 1.9 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.4 | 4.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 | 3.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.3 | 3.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.3 | 1.7 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 5.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 2.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 1.2 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 2.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 4.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 9.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 2.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 8.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 2.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.9 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 3.6 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 3.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.6 | 4.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 2.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.3 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 2.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 11.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 3.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 8.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 9.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 2.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 5.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 6.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.0 | 5.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.7 | 3.0 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.6 | 2.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.5 | 4.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 2.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 1.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 3.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 4.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 2.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.9 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 1.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 4.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 3.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 5.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 6.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 4.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 11.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 4.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 5.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |