Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
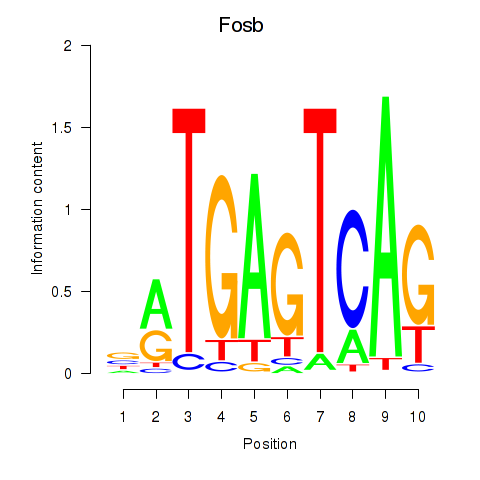
Results for Fosb
Z-value: 0.47
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSRNOG00000046667 | FosB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosb | rn6_v1_chr1_-_80221710_80221710 | -0.17 | 2.3e-03 | Click! |
Activity profile of Fosb motif
Sorted Z-values of Fosb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_81821127 | 9.78 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr15_-_77736892 | 7.98 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr1_-_199395363 | 7.42 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr20_+_3558827 | 7.31 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_149417212 | 7.21 |
ENSRNOT00000018573
|
Gpr87
|
G protein-coupled receptor 87 |
| chr1_+_20332371 | 7.06 |
ENSRNOT00000037259
|
Tmem200a
|
transmembrane protein 200A |
| chr12_-_2568382 | 6.91 |
ENSRNOT00000035142
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr2_-_178297172 | 6.14 |
ENSRNOT00000038543
|
Fnip2
|
folliculin interacting protein 2 |
| chr2_+_144861455 | 6.13 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr14_+_104250617 | 5.90 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chrX_-_142248369 | 5.83 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_-_186480278 | 5.78 |
ENSRNOT00000031743
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr1_+_221236773 | 5.57 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr1_-_31122093 | 5.19 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr2_-_28799266 | 4.91 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr4_-_157294047 | 4.88 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr19_+_52077501 | 4.85 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr9_+_20251521 | 4.66 |
ENSRNOT00000005535
|
LOC100911625
|
gamma-enolase-like |
| chr8_+_5676665 | 4.61 |
ENSRNOT00000012310
|
Mmp3
|
matrix metallopeptidase 3 |
| chr19_+_52077109 | 4.50 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr14_+_2892753 | 4.32 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr14_-_21127868 | 4.15 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr15_-_34444244 | 3.86 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr3_+_110442637 | 3.43 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr1_-_256813711 | 2.61 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr1_-_142197182 | 2.61 |
ENSRNOT00000015521
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr19_+_41631715 | 2.39 |
ENSRNOT00000022749
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr13_+_51384389 | 2.29 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr20_+_5040337 | 2.24 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr6_-_71199110 | 1.93 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chr7_+_23854846 | 1.87 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr10_-_88152064 | 1.87 |
ENSRNOT00000019477
|
Krt16
|
keratin 16 |
| chr20_-_3419831 | 1.80 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr10_-_90312386 | 1.69 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr6_-_107678156 | 1.32 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr8_+_5522739 | 1.24 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr20_+_25990304 | 1.23 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr4_-_119148358 | 1.23 |
ENSRNOT00000012218
|
Gkn1
|
gastrokine 1 |
| chr1_+_220826560 | 1.23 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr4_+_176994129 | 1.22 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr20_+_25990656 | 1.03 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_22964230 | 0.90 |
ENSRNOT00000041813
|
LOC100362149
|
ribosomal protein S20-like |
| chr1_+_218466289 | 0.67 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr8_+_5703206 | 0.65 |
ENSRNOT00000012595
|
Mmp1
|
matrix metallopeptidase 1 |
| chr13_+_90533365 | 0.51 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr10_-_71382058 | 0.44 |
ENSRNOT00000043148
|
Dusp14
|
dual specificity phosphatase 14 |
| chr2_+_204886202 | 0.41 |
ENSRNOT00000022200
|
Ngf
|
nerve growth factor |
| chr5_+_135687538 | 0.36 |
ENSRNOT00000091664
|
AC126292.2
|
|
| chr15_+_61692116 | 0.35 |
ENSRNOT00000071455
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr5_-_16706909 | 0.32 |
ENSRNOT00000011314
|
Rps20
|
ribosomal protein S20 |
| chr18_+_35249137 | 0.14 |
ENSRNOT00000017489
|
LOC100910942
|
serine protease inhibitor Kazal-type 6-like |
| chr10_-_87232723 | 0.11 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr9_-_17206994 | 0.04 |
ENSRNOT00000026195
|
Gtpbp2
|
GTP binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.1 | 9.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.9 | 2.6 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.9 | 2.6 | GO:2000645 | negative regulation of neurotrophin production(GO:0032900) nerve growth factor production(GO:0032902) negative regulation of receptor catabolic process(GO:2000645) |
| 0.8 | 5.8 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.7 | 7.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 4.6 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.6 | 7.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.6 | 2.4 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.6 | 2.3 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 1.7 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.3 | 1.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 5.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 1.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 0.6 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.2 | 2.3 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 | 4.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 6.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 3.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.9 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 5.8 | GO:0007588 | excretion(GO:0007588) |
| 0.1 | 0.4 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.1 | 4.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 6.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 9.1 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 7.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 9.8 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 3.4 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 8.0 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 4.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 9.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 13.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 5.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 7.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.1 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.0 | 7.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 6.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.8 | 5.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.7 | 5.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.6 | 7.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 6.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 2.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 2.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 2.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 2.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 5.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.1 | 1.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.7 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 5.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 10.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 6.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 4.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 9.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 5.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |