Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Fli1
Z-value: 0.87
Transcription factors associated with Fli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fli1
|
ENSRNOG00000008904 | Fli-1 proto-oncogene, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | rn6_v1_chr8_-_33661049_33661049 | 0.03 | 5.5e-01 | Click! |
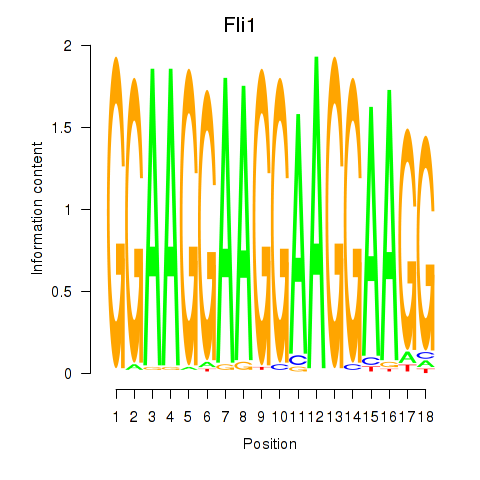
Activity profile of Fli1 motif
Sorted Z-values of Fli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_80594136 | 133.20 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr1_+_150797084 | 83.63 |
ENSRNOT00000018990
|
Nox4
|
NADPH oxidase 4 |
| chr8_-_62458301 | 68.12 |
ENSRNOT00000021653
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr8_+_128044084 | 54.17 |
ENSRNOT00000019106
|
Xylb
|
xylulokinase |
| chr20_-_9876008 | 39.83 |
ENSRNOT00000001537
|
Tff2
|
trefoil factor 2 |
| chr4_+_14039977 | 38.66 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr7_+_143754892 | 37.45 |
ENSRNOT00000085896
|
Soat2
|
sterol O-acyltransferase 2 |
| chr1_-_259287684 | 37.26 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr1_+_80383050 | 32.74 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr1_-_20155960 | 28.67 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr6_-_127653124 | 26.17 |
ENSRNOT00000047324
|
Serpina11
|
serpin family A member 11 |
| chr10_+_65733991 | 25.18 |
ENSRNOT00000013698
|
Slc46a1
|
solute carrier family 46 member 1 |
| chr7_-_67345295 | 23.79 |
ENSRNOT00000087045
ENSRNOT00000005829 |
Avpr1a
|
arginine vasopressin receptor 1A |
| chr18_-_59819113 | 22.18 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr15_-_45550285 | 21.52 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr4_-_51199570 | 19.19 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr1_+_61445672 | 17.26 |
ENSRNOT00000089298
|
LOC108348215
|
zinc finger protein 420-like |
| chr1_+_248723397 | 16.77 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr4_-_145699577 | 16.24 |
ENSRNOT00000014377
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr8_-_6203515 | 16.08 |
ENSRNOT00000087278
ENSRNOT00000031189 ENSRNOT00000008074 ENSRNOT00000085285 ENSRNOT00000007866 |
Yap1
|
yes-associated protein 1 |
| chr20_-_20680675 | 15.02 |
ENSRNOT00000087085
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr18_-_80865584 | 15.01 |
ENSRNOT00000021750
|
Tshz1
|
teashirt zinc finger homeobox 1 |
| chr7_+_23913830 | 14.81 |
ENSRNOT00000006570
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr8_+_37156619 | 12.73 |
ENSRNOT00000070856
|
Gm17689
|
predicted gene, 17689 |
| chr7_-_2623781 | 12.31 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr2_+_196608496 | 11.37 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr7_-_70926903 | 11.26 |
ENSRNOT00000031005
|
Lrp1
|
LDL receptor related protein 1 |
| chr9_-_17212628 | 10.84 |
ENSRNOT00000002656
ENSRNOT00000007876 |
RGD1562399
|
similar to 40S ribosomal protein S2 |
| chr13_-_111972603 | 10.47 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr4_-_82229397 | 10.24 |
ENSRNOT00000089432
|
Hoxa13
|
homeo box A13 |
| chrX_+_159560292 | 10.09 |
ENSRNOT00000087250
|
Vgll1
|
vestigial like family member 1 |
| chr14_-_84189266 | 9.93 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr2_+_248219428 | 9.88 |
ENSRNOT00000037181
|
LOC685067
|
similar to guanylate binding protein family, member 6 |
| chr12_+_41636994 | 9.77 |
ENSRNOT00000001882
|
Sdsl
|
serine dehydratase-like |
| chr4_-_163528220 | 9.76 |
ENSRNOT00000090813
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr3_+_147992754 | 9.10 |
ENSRNOT00000055412
|
Defb28
|
defensin beta 28 |
| chr1_-_64021321 | 8.94 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr3_-_176744377 | 8.49 |
ENSRNOT00000017787
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr18_+_63098144 | 8.41 |
ENSRNOT00000024968
|
Cidea
|
cell death-inducing DFFA-like effector a |
| chr4_-_158705885 | 8.07 |
ENSRNOT00000087115
ENSRNOT00000026685 |
Ntf3
|
neurotrophin 3 |
| chrX_+_80213332 | 7.33 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr18_-_55992885 | 7.02 |
ENSRNOT00000025881
|
Ndst1
|
N-deacetylase and N-sulfotransferase 1 |
| chr5_+_164751440 | 6.94 |
ENSRNOT00000010558
|
RGD1305350
|
similar to RIKEN cDNA 2510039O18 |
| chr4_-_156426760 | 6.73 |
ENSRNOT00000014028
|
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr3_-_103345333 | 6.54 |
ENSRNOT00000071479
|
LOC100909692
|
olfactory receptor 4F3/4F16/4F29-like |
| chr5_+_173024335 | 6.52 |
ENSRNOT00000023097
|
Slc35e2b
|
solute carrier family 35, member E2B |
| chr16_+_7758996 | 6.28 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr15_-_34322115 | 5.78 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr17_+_57074525 | 5.60 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr12_+_25264192 | 5.51 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr12_+_25264400 | 5.50 |
ENSRNOT00000087656
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr15_-_35166211 | 5.45 |
ENSRNOT00000059662
|
LOC691670
|
similar to natural killer cell protease 7 |
| chr17_-_14828329 | 5.43 |
ENSRNOT00000022830
|
Barx1
|
BARX homeobox 1 |
| chr3_+_4106477 | 5.21 |
ENSRNOT00000078985
|
AABR07051251.1
|
|
| chrX_+_15378789 | 5.17 |
ENSRNOT00000029272
|
Gata1
|
GATA binding protein 1 |
| chr1_+_264504591 | 5.16 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr3_-_67668772 | 5.09 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr3_+_33641616 | 5.06 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr4_+_156050054 | 4.79 |
ENSRNOT00000039855
|
Clec4a
|
C-type lectin domain family 4, member A |
| chr7_+_25919867 | 4.78 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr10_-_11174861 | 4.78 |
ENSRNOT00000006680
|
Glis2
|
GLIS family zinc finger 2 |
| chr8_+_48805684 | 4.76 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr1_+_225690756 | 4.66 |
ENSRNOT00000027498
|
Psbpc2
|
prostatic steroid-binding protein C2 |
| chr20_+_47596575 | 4.54 |
ENSRNOT00000087230
ENSRNOT00000064905 |
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr19_-_39087880 | 4.50 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr3_-_7203420 | 4.49 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr10_+_4538111 | 4.46 |
ENSRNOT00000084151
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr5_-_135628750 | 4.44 |
ENSRNOT00000079236
|
AC126292.3
|
|
| chr3_+_5555807 | 4.43 |
ENSRNOT00000007677
|
Cacfd1
|
calcium channel flower domain containing 1 |
| chr20_-_32353677 | 4.38 |
ENSRNOT00000035355
|
Stox1
|
storkhead box 1 |
| chr8_+_111239922 | 4.36 |
ENSRNOT00000077948
|
AABR07071358.1
|
|
| chr4_+_120843453 | 4.35 |
ENSRNOT00000022534
|
Tpra1
|
transmembrane protein adipocyte associated 1 |
| chr19_+_38619764 | 4.29 |
ENSRNOT00000027225
|
LOC100360573
|
ribosomal protein S12-like |
| chr12_-_23841049 | 4.29 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chrX_+_156355376 | 4.28 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr2_+_250600823 | 4.24 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
| chr10_-_44256945 | 4.20 |
ENSRNOT00000003818
|
Olr1432
|
olfactory receptor 1432 |
| chr15_-_20783063 | 4.19 |
ENSRNOT00000083268
|
Bmp4
|
bone morphogenetic protein 4 |
| chr10_-_65502936 | 4.17 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr5_+_64566804 | 4.00 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr10_+_67325347 | 3.95 |
ENSRNOT00000079002
ENSRNOT00000085914 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chrX_+_157759624 | 3.81 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chrX_+_43625169 | 3.80 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr5_-_150390099 | 3.79 |
ENSRNOT00000014479
|
Ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr9_-_50884596 | 3.77 |
ENSRNOT00000016285
|
Kdelc1
|
KDEL motif containing 1 |
| chr2_+_29410579 | 3.76 |
ENSRNOT00000023180
|
Zfp366
|
zinc finger protein 366 |
| chr1_+_190200324 | 3.65 |
ENSRNOT00000001933
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr3_-_103400272 | 3.62 |
ENSRNOT00000046527
|
Olr790
|
olfactory receptor 790 |
| chr14_-_86796378 | 3.48 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr20_+_12773427 | 3.45 |
ENSRNOT00000001695
|
Col6a2
|
collagen type VI alpha 2 chain |
| chr3_+_66593934 | 3.42 |
ENSRNOT00000008046
|
Ssfa2
|
sperm specific antigen 2 |
| chr18_+_55666027 | 3.36 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr1_+_256370850 | 3.28 |
ENSRNOT00000030962
|
Cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr5_-_147907733 | 2.99 |
ENSRNOT00000032558
|
Tmem39b
|
transmembrane protein 39b |
| chr1_+_99599620 | 2.90 |
ENSRNOT00000024690
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr5_+_173237642 | 2.82 |
ENSRNOT00000068770
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr20_+_48881194 | 2.68 |
ENSRNOT00000000304
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr5_+_57738321 | 2.66 |
ENSRNOT00000063849
|
Ubap1
|
ubiquitin-associated protein 1 |
| chr2_+_154604832 | 2.62 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr18_+_57286322 | 2.56 |
ENSRNOT00000026174
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr20_+_18492169 | 2.54 |
ENSRNOT00000046536
|
LOC103690821
|
60S ribosomal protein L39 |
| chr16_-_69242028 | 2.54 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr4_-_170932618 | 2.50 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr8_-_79862974 | 2.47 |
ENSRNOT00000091386
|
AC128582.1
|
|
| chr7_-_140546908 | 2.41 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr16_+_20020444 | 2.35 |
ENSRNOT00000024827
|
Pgls
|
6-phosphogluconolactonase |
| chr7_-_116010938 | 2.35 |
ENSRNOT00000076807
ENSRNOT00000076047 |
D730001G18Rik
|
RIKEN cDNA D730001G18 gene |
| chrX_-_4957529 | 2.20 |
ENSRNOT00000000173
|
Dusp21
|
dual specificity phosphatase 21 |
| chr7_+_137680530 | 2.20 |
ENSRNOT00000006970
|
Arid2
|
AT-rich interaction domain 2 |
| chrX_-_113474164 | 2.19 |
ENSRNOT00000046804
|
Gucy2f
|
guanylate cyclase 2F |
| chr8_+_20230082 | 2.16 |
ENSRNOT00000044463
|
Olr1165
|
olfactory receptor 1165 |
| chr10_+_45893018 | 2.14 |
ENSRNOT00000004280
ENSRNOT00000086710 |
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr4_-_156563438 | 2.12 |
ENSRNOT00000045305
|
Vom2r51
|
vomeronasal 2 receptor, 51 |
| chr10_-_13580821 | 2.00 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr15_-_30019055 | 1.83 |
ENSRNOT00000073984
|
AABR07017681.1
|
|
| chr10_+_46840113 | 1.82 |
ENSRNOT00000086083
ENSRNOT00000079133 |
Myo15a
|
myosin XVA |
| chr1_-_24302298 | 1.81 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_24601063 | 1.78 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr16_-_69884962 | 1.65 |
ENSRNOT00000078635
|
Cnga2
|
cyclic nucleotide gated channel alpha 2 |
| chr9_+_81427730 | 1.64 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chrX_+_158569056 | 1.63 |
ENSRNOT00000074170
|
LOC102550080
|
placenta-specific protein 1-like |
| chr19_+_39087990 | 1.63 |
ENSRNOT00000027553
|
Utp4
|
UTP4 small subunit processome component |
| chr12_-_17186679 | 1.58 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr8_+_49354115 | 1.56 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr5_+_159534202 | 1.56 |
ENSRNOT00000010993
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr13_-_72869396 | 1.48 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
| chr20_-_32080088 | 1.46 |
ENSRNOT00000080773
|
Supv3l1
|
Suv3 like RNA helicase |
| chr1_-_85317968 | 1.40 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr8_+_79159370 | 1.39 |
ENSRNOT00000086841
|
Rfx7
|
regulatory factor X, 7 |
| chr2_-_250600517 | 1.39 |
ENSRNOT00000016872
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr16_+_35934970 | 1.36 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr17_+_6684621 | 1.36 |
ENSRNOT00000089138
|
RGD1311345
|
similar to CG9752-PA |
| chr6_-_111572949 | 1.32 |
ENSRNOT00000016884
|
RGD1565693
|
similar to GLE1-like, RNA export mediator isoform 1 |
| chr10_-_61577412 | 1.28 |
ENSRNOT00000003696
|
Pafah1b1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 |
| chr12_+_12756452 | 1.21 |
ENSRNOT00000001382
ENSRNOT00000092462 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr5_+_137257637 | 1.17 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr5_+_151573092 | 1.17 |
ENSRNOT00000011049
|
Slc9a1
|
solute carrier family 9 member A1 |
| chr14_-_45863877 | 1.15 |
ENSRNOT00000002977
|
Pgm2
|
phosphoglucomutase 2 |
| chr1_+_45923222 | 1.07 |
ENSRNOT00000092976
ENSRNOT00000084454 ENSRNOT00000022939 |
Arid1b
|
AT-rich interaction domain 1B |
| chr3_-_33075685 | 1.05 |
ENSRNOT00000006937
|
Orc4
|
origin recognition complex, subunit 4 |
| chr20_-_6556350 | 0.97 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chr7_+_107467260 | 0.94 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr2_-_27287605 | 0.77 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr3_+_43255567 | 0.71 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr18_-_49937522 | 0.63 |
ENSRNOT00000033254
|
Zfp608
|
zinc finger protein 608 |
| chr15_-_29246222 | 0.60 |
ENSRNOT00000081806
|
AABR07017617.1
|
|
| chr11_+_86852711 | 0.54 |
ENSRNOT00000002581
|
Dgcr8
|
DGCR8 microprocessor complex subunit |
| chr9_-_24383679 | 0.53 |
ENSRNOT00000040542
|
Crisp1
|
cysteine-rich secretory protein 1 |
| chr2_+_266315036 | 0.50 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr5_+_117877594 | 0.47 |
ENSRNOT00000011631
|
Atg4c
|
autophagy related 4C, cysteine peptidase |
| chr7_-_13425311 | 0.42 |
ENSRNOT00000060455
|
Olr1076
|
olfactory receptor 1076 |
| chr1_+_217699180 | 0.40 |
ENSRNOT00000042579
|
LOC100364769
|
LRRG00136-like |
| chr11_-_65209268 | 0.29 |
ENSRNOT00000077612
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr14_+_100415668 | 0.27 |
ENSRNOT00000008057
|
C1d
|
C1D nuclear receptor co-repressor |
| chr20_-_11528332 | 0.27 |
ENSRNOT00000038419
|
Tspear
|
thrombospondin-type laminin G domain and EAR repeats |
| chr4_+_78735279 | 0.21 |
ENSRNOT00000011970
|
Malsu1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr14_+_70780623 | 0.16 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr2_-_230356919 | 0.12 |
ENSRNOT00000038509
|
Sec24b
|
SEC24 homolog B, COPII coat complex component |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.4 | 133.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 18.1 | 54.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 13.6 | 68.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 7.9 | 23.8 | GO:0042631 | cellular response to water deprivation(GO:0042631) positive regulation of glycogen catabolic process(GO:0045819) |
| 6.4 | 38.7 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 6.3 | 25.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 5.7 | 39.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 4.6 | 83.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 4.2 | 37.5 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 3.5 | 10.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 3.4 | 10.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 3.0 | 9.0 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 2.8 | 11.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 2.7 | 16.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 2.5 | 15.0 | GO:0060023 | soft palate development(GO:0060023) |
| 2.2 | 6.7 | GO:1901090 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 2.2 | 8.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 2.0 | 8.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 2.0 | 9.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.8 | 7.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 1.7 | 5.2 | GO:0090202 | regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 1.7 | 5.2 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 1.7 | 5.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 1.6 | 11.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.6 | 37.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.4 | 4.3 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 1.4 | 1.4 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 1.3 | 14.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 1.2 | 8.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.2 | 16.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.1 | 19.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.1 | 5.6 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 1.0 | 3.8 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.9 | 3.8 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.8 | 2.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.8 | 3.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.8 | 11.4 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.6 | 4.5 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.6 | 4.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.6 | 2.9 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.5 | 2.1 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.5 | 4.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 3.5 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.5 | 2.4 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 23.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.4 | 16.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.4 | 4.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 1.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.4 | 4.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 2.3 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.3 | 1.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 | 1.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.3 | 4.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 2.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 2.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 1.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.2 | 1.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.2 | 1.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 3.8 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 6.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 1.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 2.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 1.2 | GO:0035338 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 3.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 5.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.7 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 2.3 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 4.8 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 5.4 | GO:0048536 | spleen development(GO:0048536) |
| 0.1 | 0.3 | GO:0071282 | cellular response to iron(II) ion(GO:0071282) |
| 0.1 | 25.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.5 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 5.8 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 4.9 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 4.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 14.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.2 | 133.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 8.4 | 83.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 5.4 | 16.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 1.9 | 14.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.8 | 16.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.3 | 5.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.9 | 23.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.7 | 63.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.6 | 4.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.5 | 4.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 1.5 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.5 | 4.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 37.5 | GO:0005903 | brush border(GO:0005903) |
| 0.3 | 6.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 2.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.3 | 2.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 2.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 20.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 5.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 4.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 5.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 8.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 10.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 113.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 3.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 3.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 11.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 11.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 23.9 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 1.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 6.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 5.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 7.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 63.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.5 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.3 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.4 | 133.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 9.9 | 88.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 7.9 | 23.8 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 7.7 | 38.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 6.3 | 25.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 4.9 | 68.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 3.8 | 11.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 3.5 | 10.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.8 | 39.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.1 | 6.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.8 | 8.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.6 | 54.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 1.4 | 4.2 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 1.3 | 14.8 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 1.3 | 6.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.3 | 37.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 1.3 | 37.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.3 | 3.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.2 | 9.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.2 | 19.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 1.2 | 7.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.1 | 3.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.0 | 8.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.9 | 23.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 1.6 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.5 | 11.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.5 | 16.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 4.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 3.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.5 | 4.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 4.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 5.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 1.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 1.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 1.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 0.7 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 1.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 26.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 2.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 4.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 1.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 3.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 4.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 5.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 5.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 15.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 8.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.3 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 2.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 2.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.5 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.3 | GO:0044212 | regulatory region DNA binding(GO:0000975) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 25.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 13.1 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.6 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 14.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 83.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.5 | 11.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.5 | 16.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 11.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 8.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 5.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 5.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 24.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 4.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.7 | 68.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 7.8 | 133.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.4 | 19.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.0 | 16.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.7 | 16.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 4.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 36.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.5 | 11.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.4 | 25.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 10.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 8.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 3.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 3.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 8.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 23.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 16.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.1 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.1 | 3.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 5.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.3 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |