Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Figla
Z-value: 0.89
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSRNOG00000015877 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Figla | rn6_v1_chr4_+_117679342_117679342 | 0.06 | 2.9e-01 | Click! |
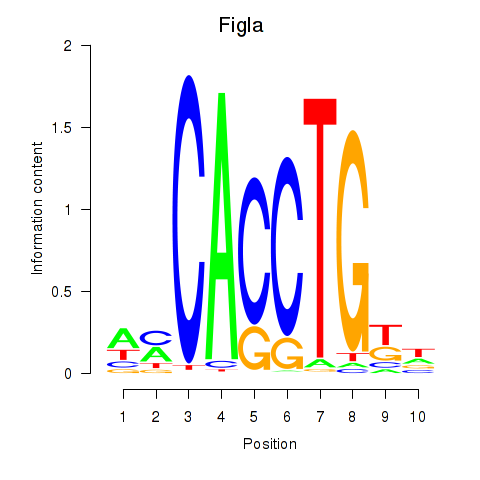
Activity profile of Figla motif
Sorted Z-values of Figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87774552 | 98.63 |
ENSRNOT00000044342
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr10_+_87782376 | 44.61 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chr10_-_87564327 | 41.32 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr2_-_105089659 | 35.89 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr10_+_87759769 | 35.18 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr5_-_155772040 | 29.26 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr4_-_28437676 | 23.27 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr10_+_87788458 | 19.74 |
ENSRNOT00000042020
|
LOC100910814
|
keratin-associated protein 9-1-like |
| chr10_-_87529599 | 19.32 |
ENSRNOT00000074099
|
Krtap2-1
|
keratin associated protein 2-1 |
| chr7_-_143016040 | 18.60 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr4_+_56981283 | 17.57 |
ENSRNOT00000010989
|
Tspan33
|
tetraspanin 33 |
| chr5_-_24631679 | 17.44 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr15_-_37383277 | 16.77 |
ENSRNOT00000011711
|
Gjb2
|
gap junction protein, beta 2 |
| chr10_+_104368247 | 16.73 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr7_-_71226150 | 14.98 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr5_+_154522119 | 14.85 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr10_+_103713045 | 13.93 |
ENSRNOT00000004351
|
Slc9a3r1
|
SLC9A3 regulator 1 |
| chr3_+_112531703 | 12.23 |
ENSRNOT00000041727
|
LOC100911204
|
protein CASC5-like |
| chr10_+_106812739 | 11.58 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr4_-_69196430 | 11.20 |
ENSRNOT00000017673
|
LOC312273
|
Trypsin V-A |
| chr18_+_61377051 | 11.00 |
ENSRNOT00000066659
|
Oacyl
|
O-acyltransferase like |
| chr2_+_77868412 | 10.86 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr10_-_108196217 | 10.75 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr1_+_80000165 | 10.60 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr12_+_9728486 | 10.36 |
ENSRNOT00000001263
|
Lnx2
|
ligand of numb-protein X 2 |
| chr4_-_82271893 | 10.16 |
ENSRNOT00000075005
|
Hoxa7
|
homeobox A7 |
| chrX_-_154918095 | 10.09 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr7_-_101140308 | 9.82 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr5_-_169212170 | 9.42 |
ENSRNOT00000013385
|
Tas1r1
|
taste 1 receptor member 1 |
| chr8_+_62341613 | 9.26 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr1_+_100470722 | 9.19 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr11_-_90406797 | 8.63 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr3_+_141927400 | 8.26 |
ENSRNOT00000066588
|
AABR07054117.1
|
|
| chr4_-_82202096 | 7.87 |
ENSRNOT00000081824
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr1_-_89269930 | 7.84 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr9_-_18371856 | 7.78 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr10_-_87541851 | 7.67 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr1_+_224824799 | 7.61 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr1_+_41325462 | 7.60 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr2_-_43068677 | 7.47 |
ENSRNOT00000036658
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr15_+_32355565 | 7.19 |
ENSRNOT00000072382
|
AABR07017900.1
|
|
| chr3_+_161236898 | 7.01 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_-_80311043 | 7.01 |
ENSRNOT00000068092
|
Klc3
|
kinesin light chain 3 |
| chr9_+_66335492 | 6.53 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr6_+_26051396 | 6.27 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr1_+_7252349 | 5.84 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr3_+_125503638 | 5.52 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr10_-_87578854 | 5.43 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr1_-_221431713 | 5.31 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr16_-_39970532 | 5.26 |
ENSRNOT00000071331
|
Spata4
|
spermatogenesis associated 4 |
| chr1_+_78739930 | 5.18 |
ENSRNOT00000021976
|
Strn4
|
striatin 4 |
| chr17_-_8619737 | 5.12 |
ENSRNOT00000065217
|
RGD1562024
|
RGD1562024 |
| chr10_-_89088993 | 5.09 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr10_-_70220558 | 4.97 |
ENSRNOT00000041389
ENSRNOT00000076398 ENSRNOT00000076596 |
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr13_-_110642754 | 4.92 |
ENSRNOT00000029039
|
LOC100362350
|
hydroxysteroid 17-beta dehydrogenase 6-like |
| chr10_-_87521514 | 4.82 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr10_-_87535438 | 4.75 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr3_+_148386189 | 4.67 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr9_+_88357556 | 4.53 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr4_-_82186190 | 4.35 |
ENSRNOT00000071729
|
LOC100911622
|
homeobox protein Hox-A7-like |
| chr8_+_116754178 | 4.32 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_-_87753668 | 4.29 |
ENSRNOT00000066758
ENSRNOT00000047104 |
Krtap4-3
|
keratin associated protein 4-3 |
| chr15_+_4475051 | 4.23 |
ENSRNOT00000008461
|
Kcnk16
|
potassium two pore domain channel subfamily K member 16 |
| chr1_+_277355619 | 4.07 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr20_-_2701637 | 3.95 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr20_-_2210033 | 3.78 |
ENSRNOT00000084596
|
Trim26
|
tripartite motif-containing 26 |
| chr20_-_4879779 | 3.51 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr12_-_46414434 | 3.50 |
ENSRNOT00000041281
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr18_+_48132414 | 3.33 |
ENSRNOT00000050631
|
Snx2
|
sorting nexin 2 |
| chr10_-_56289882 | 3.30 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr17_-_42267079 | 3.29 |
ENSRNOT00000059564
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr20_-_5865775 | 3.20 |
ENSRNOT00000000612
ENSRNOT00000092641 |
Srpk1
|
SRSF protein kinase 1 |
| chr10_+_87832743 | 3.19 |
ENSRNOT00000055286
|
Krtap31-1
|
keratin associated protein 31-1 |
| chr3_-_152179193 | 3.17 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr10_-_54467956 | 3.12 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr3_+_7686503 | 3.11 |
ENSRNOT00000017984
|
Setx
|
senataxin |
| chr1_+_192025710 | 3.08 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_-_192025350 | 2.98 |
ENSRNOT00000071946
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr11_+_50781127 | 2.95 |
ENSRNOT00000002738
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr12_-_47360507 | 2.77 |
ENSRNOT00000001562
|
Sppl3
|
signal peptide peptidase like 3 |
| chr3_+_103753238 | 2.63 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr8_-_22874637 | 2.50 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr13_-_73558912 | 2.47 |
ENSRNOT00000093655
|
Cep350
|
centrosomal protein 350 |
| chr10_+_87846947 | 2.45 |
ENSRNOT00000077436
|
Krtap9-5
|
keratin associated protein 9-5 |
| chr4_-_129846642 | 2.45 |
ENSRNOT00000010717
|
Frmd4b
|
FERM domain containing 4B |
| chr2_+_95008477 | 2.44 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr4_+_127164453 | 2.40 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr1_-_215423099 | 2.35 |
ENSRNOT00000027259
|
AABR07006049.1
|
|
| chr20_+_14101659 | 2.35 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr10_+_16542882 | 2.31 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chrX_-_111942749 | 2.20 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr8_+_111210811 | 2.12 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr2_+_95008311 | 2.03 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr10_+_65772443 | 1.96 |
ENSRNOT00000013296
|
Sebox
|
SEBOX homeobox |
| chr10_+_87808493 | 1.92 |
ENSRNOT00000065405
|
LOC680428
|
hypothetical protein LOC680428 |
| chr2_+_197720259 | 1.80 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr1_+_82151669 | 1.77 |
ENSRNOT00000091357
|
Cic
|
capicua transcriptional repressor |
| chr11_-_35749464 | 1.60 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr7_-_2912491 | 1.56 |
ENSRNOT00000048453
|
AC128207.1
|
|
| chr6_+_75996643 | 1.56 |
ENSRNOT00000052076
|
Srp54a
|
signal recognition particle 54A |
| chr9_+_51009116 | 1.55 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr19_-_55434252 | 1.51 |
ENSRNOT00000045052
|
Pabpn1l
|
poly(A)binding protein nuclear 1-like |
| chr7_-_143552588 | 1.48 |
ENSRNOT00000086317
ENSRNOT00000092619 ENSRNOT00000092138 |
Krt78
|
keratin 78 |
| chr6_+_50528823 | 1.37 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr8_-_50126413 | 1.22 |
ENSRNOT00000072712
ENSRNOT00000022583 |
Cep164
|
centrosomal protein 164 |
| chr3_+_11476883 | 1.04 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr3_+_73089269 | 0.93 |
ENSRNOT00000084321
|
Olr453
|
olfactory receptor 453 |
| chr19_+_22699808 | 0.78 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr4_+_88048267 | 0.77 |
ENSRNOT00000044913
|
Vom1r81
|
vomeronasal 1 receptor 81 |
| chr4_-_68597586 | 0.59 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr18_+_3887419 | 0.50 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr3_+_140024043 | 0.42 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr10_-_43895376 | 0.12 |
ENSRNOT00000031811
|
Olr1413
|
olfactory receptor 1413 |
| chr3_-_60460724 | 0.04 |
ENSRNOT00000024706
|
Chrna1
|
cholinergic receptor nicotinic alpha 1 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 4.2 | 16.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 3.1 | 9.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 2.9 | 8.6 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.5 | 7.5 | GO:1902380 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 2.5 | 14.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 2.1 | 6.3 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 2.0 | 7.8 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 1.9 | 7.6 | GO:1990375 | baculum development(GO:1990375) |
| 1.8 | 7.0 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 1.6 | 9.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 1.6 | 4.7 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 1.5 | 7.6 | GO:0031427 | response to methotrexate(GO:0031427) |
| 1.3 | 16.8 | GO:0016264 | gap junction assembly(GO:0016264) cellular response to glucagon stimulus(GO:0071377) |
| 1.1 | 10.2 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 1.1 | 3.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.9 | 10.6 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.9 | 2.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.9 | 4.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.8 | 5.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.8 | 5.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.8 | 3.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.6 | 3.0 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.5 | 1.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 13.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.4 | 7.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 4.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.3 | 1.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 2.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.3 | 34.6 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.2 | 2.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 3.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.8 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 10.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 3.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 10.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 3.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 2.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 9.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.9 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 2.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 3.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 5.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 11.2 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 4.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 14.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 4.3 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 4.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.0 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 3.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 10.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.0 | GO:0048477 | oogenesis(GO:0048477) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.7 | 129.9 | GO:0045095 | keratin filament(GO:0045095) |
| 1.2 | 5.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 1.0 | 13.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 16.8 | GO:0097449 | connexon complex(GO:0005922) astrocyte projection(GO:0097449) |
| 0.8 | 7.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.7 | 10.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.6 | 10.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.6 | 7.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.6 | 4.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.5 | 7.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 2.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 7.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 1.4 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.3 | 9.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 3.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 23.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 11.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.5 | GO:0005608 | laminin-3 complex(GO:0005608) laminin-5 complex(GO:0005610) |
| 0.1 | 5.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 7.6 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 3.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 5.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 10.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 14.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 23.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 8.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 32.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 3.5 | 13.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 2.5 | 7.5 | GO:0031249 | denatured protein binding(GO:0031249) |
| 1.5 | 7.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.3 | 35.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.2 | 16.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 1.2 | 4.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.1 | 3.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.0 | 7.6 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.7 | 2.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.6 | 10.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.5 | 5.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 2.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.4 | 9.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 4.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.4 | 1.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 5.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 9.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 9.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.3 | 5.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 7.0 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.2 | 5.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 30.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 6.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 40.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 7.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 7.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 5.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 9.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 22.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 14.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 17.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 8.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 3.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 2.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 5.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 94.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 3.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 12.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 3.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 3.6 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 3.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 10.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 7.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 10.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 13.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 8.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 6.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 27.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 7.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 3.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.9 | 14.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.7 | 7.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 7.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.4 | 9.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 5.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.3 | 4.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 5.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 7.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 10.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 7.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 7.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 4.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 6.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 4.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 4.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |